Imaging mass spectrometry of gastric carcinoma in formalin-fixed paraffin-embedded tissue microarray
- PMID: 19961487
- PMCID: PMC11158157
- DOI: 10.1111/j.1349-7006.2009.01384.x
Imaging mass spectrometry of gastric carcinoma in formalin-fixed paraffin-embedded tissue microarray
Abstract
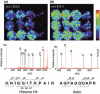
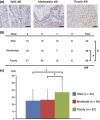
The popularity of imaging mass spectrometry (IMS) of tissue samples, which enables the direct scanning of tissue sections within a short time-period, has been considerably increasing in cancer proteomics. Most pathological specimens stored in medical institutes are formalin-fixed; thus, they had been regarded to be unsuitable for proteomic analyses, including IMS, until recently. Here, we report an easy-to-use screening method that enables the analysis of multiple samples in one experiment without extractions and purifications of proteins. We scanned, with an IMS technique, a tissue microarray (TMA) of formalin-fixed paraffin-embedded (FFPE) specimens. We detected a large amount of signals from trypsin-treated FFPE-TMA samples of gastric carcinoma tissues of different histological types. Of the signals detected, 54 were classified as signals specific to cancer with statistically significant differences between adenocarcinomas and normal tissues. We detected a total of 14 of the 54 signals as histological type-specific with the support of statistical analyses. Tandem MS revealed that a signal specific to poorly differentiated cancer tissue corresponded to histone H4. Finally, we verified the IMS-based finding by immunohistochemical analysis of more than 300 specimens spotted on TMAs; the immunoreactivity of histone H4 was remarkably strong in poorly differentiated cancer tissues. Thus, the application of IMS to FFPE-TMA can enable high-throughput analysis in cancer proteomics to aid in the understanding of molecular mechanisms underlying carcinogenesis, invasiveness, metastasis, and prognosis. Further, results obtained from the IMS of FFPE-TMA can be readily confirmed by commonly used immunohistochemical analyses.
Figures
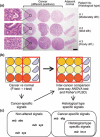
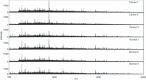

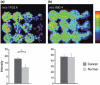
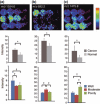
References
-
- DeRisi J, Penland L, Brown PO et al. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet 1996. Dec; 14 (4): 457–60. - PubMed
-
- Ramsay G. DNA chips: state‐of‐the art. Nat Biotechnol 1998. Jan; 16 (1): 40–4. - PubMed
-
- Zhao X, Li C, Paez JG et al. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res 2004. May 1; 64 (9): 3060–71. - PubMed
-
- Engle LJ, Simpson CL, Landers JE. Using high‐throughput SNP technologies to study cancer. Oncogene 2006. Mar 13; 25 (11): 1594–601. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

