Power of multifactor dimensionality reduction and penalized logistic regression for detecting gene-gene interaction in a case-control study
- PMID: 19961594
- PMCID: PMC2800840
- DOI: 10.1186/1471-2350-10-127
Power of multifactor dimensionality reduction and penalized logistic regression for detecting gene-gene interaction in a case-control study
Abstract
Background: There is a growing awareness that interaction between multiple genes play an important role in the risk of common, complex multi-factorial diseases. Many common diseases are affected by certain genotype combinations (associated with some genes and their interactions). The identification and characterization of these susceptibility genes and gene-gene interaction have been limited by small sample size and large number of potential interactions between genes. Several methods have been proposed to detect gene-gene interaction in a case control study. The penalized logistic regression (PLR), a variant of logistic regression with L2 regularization, is a parametric approach to detect gene-gene interaction. On the other hand, the Multifactor Dimensionality Reduction (MDR) is a nonparametric and genetic model-free approach to detect genotype combinations associated with disease risk.
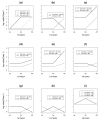
Methods: We compared the power of MDR and PLR for detecting two-way and three-way interactions in a case-control study through extensive simulations. We generated several interaction models with different magnitudes of interaction effect. For each model, we simulated 100 datasets, each with 200 cases and 200 controls and 20 SNPs. We considered a wide variety of models such as models with just main effects, models with only interaction effects or models with both main and interaction effects. We also compared the performance of MDR and PLR to detect gene-gene interaction associated with acute rejection(AR) in kidney transplant patients.
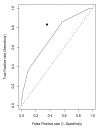
Results: In this paper, we have studied the power of MDR and PLR for detecting gene-gene interaction in a case-control study through extensive simulation. We have compared their performances for different two-way and three-way interaction models. We have studied the effect of different allele frequencies on these methods. We have also implemented their performance on a real dataset. As expected, none of these methods were consistently better for all data scenarios, but, generally MDR outperformed PLR for more complex models. The ROC analysis on the real dataset suggests that MDR outperforms PLR in detecting gene-gene interaction on the real dataset.
Conclusion: As one might expect, the relative success of each method is context dependent. This study demonstrates the strengths and weaknesses of the methods to detect gene-gene interaction.
Figures




References
-
- Hollander WF. Epistasis and hypostasis. J Hered. 1955;46:222–225.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

