Development and implementation of high-throughput SNP genotyping in barley
- PMID: 19961604
- PMCID: PMC2797026
- DOI: 10.1186/1471-2164-10-582
Development and implementation of high-throughput SNP genotyping in barley
Abstract
Background: High density genetic maps of plants have, nearly without exception, made use of marker datasets containing missing or questionable genotype calls derived from a variety of genic and non-genic or anonymous markers, and been presented as a single linear order of genetic loci for each linkage group. The consequences of missing or erroneous data include falsely separated markers, expansion of cM distances and incorrect marker order. These imperfections are amplified in consensus maps and problematic when fine resolution is critical including comparative genome analyses and map-based cloning. Here we provide a new paradigm, a high-density consensus genetic map of barley based only on complete and error-free datasets and genic markers, represented accurately by graphs and approximately by a best-fit linear order, and supported by a readily available SNP genotyping resource.
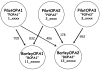
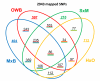
Results: Approximately 22,000 SNPs were identified from barley ESTs and sequenced amplicons; 4,596 of them were tested for performance in three pilot phase Illumina GoldenGate assays. Data from three barley doubled haploid mapping populations supported the production of an initial consensus map. Over 200 germplasm selections, principally European and US breeding material, were used to estimate minor allele frequency (MAF) for each SNP. We selected 3,072 of these tested SNPs based on technical performance, map location, MAF and biological interest to fill two 1536-SNP "production" assays (BOPA1 and BOPA2), which were made available to the barley genetics community. Data were added using BOPA1 from a fourth mapping population to yield a consensus map containing 2,943 SNP loci in 975 marker bins covering a genetic distance of 1099 cM.
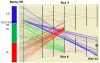
Conclusion: The unprecedented density of genic markers and marker bins enabled a high resolution comparison of the genomes of barley and rice. Low recombination in pericentric regions is evident from bins containing many more than the average number of markers, meaning that a large number of genes are recombinationally locked into the genetic centromeric regions of several barley chromosomes. Examination of US breeding germplasm illustrated the usefulness of BOPA1 and BOPA2 in that they provide excellent marker density and sensitivity for detection of minor alleles in this genetically narrow material.
Figures

References
-
- Rostoks N, Mudie S, Cardle L, Russell J, Ramsay L, Booth A, Svensson JT, Wanamaker SI, Walia H, Rodriguez EM, Hedley PE, Liu H, Morris J, Close TJ, Marshall DF, Robbie Waugh R. Genome-wide SNP discovery and linkage analysis in barley based on genes responsive to abiotic stress. Molecular Genetics and Genomics. 2005;274:515–527. doi: 10.1007/s00438-005-0046-z. - DOI - PubMed
-
- Wenzl P, Li H, Carling J, Zhou M, Raman H, Paul E, Hearnden P, Maier C, Xia L, Caig V, Ovesná J, Cakir M, Poulsen D, Wang J, Raman R, Smith KP, Muehlbauer GJ, Chalmers KJ, Kleinhofs A, Huttner E, Kilian A. A high-density consensus map of barley linking DArT markers to SSR, RFLP and STS loci and agricultural traits. BioMed Central Genomics. 2006;7:206. - PMC - PubMed
-
- Marcel TC, Varshney RK, Barbieri M, Jafary H, de Kock MJD, Graner A, Niks RE. high-density consensus map of barley to compare the distribution of QTLs for partial resistance of Puccinia hordei A and of defence gene homologues. Theoretical and Applied Genetics. 2007;114:487–500. doi: 10.1007/s00122-006-0448-2. - DOI - PubMed
-
- Stein N, Prasad M, Scholz U, Thiel T, Zhang H, Wolf M, Kota R, Varshney RK, Perovic D, Grosse I, Graner A. A 1,000-loci transcript map of the barley genome: new anchoring points for integrative grass genomics. Theoretical and Applied Genetics. 2007;114:823–839. doi: 10.1007/s00122-006-0480-2. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

