Molecular characterization and phylogenetic analysis of the complete genome of a porcine sapovirus from Chinese swine
- PMID: 19961620
- PMCID: PMC2795755
- DOI: 10.1186/1743-422X-6-216
Molecular characterization and phylogenetic analysis of the complete genome of a porcine sapovirus from Chinese swine
Abstract
Background: Porcine sapovirus was first identified in the United States in 1980, hitherto, several Asian countries have detected this virus. In 2008, the first outbreak of gastroenteritis in piglets caused by porcine sapovirus in China was reported. The complete genome of the identified SaV strain Ch-sw-sav1 was sequenced and analyzed to provide gene profile for this outbreak.
Methods: The whole genome of Ch-sw-sav1 was amplified by RT-PCR and was sequenced. Sequence alignment of the complete genome or RNA dependent RNA polymerase (RdRp) gene was done. 3' end of ORF2 with 21-nt nucleotide insertion was further analyzed using software.
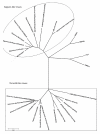
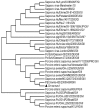
Results: Sequence analysis indicated that the genome of Ch-sw-sav1 was 7541 nucleotide long with two ORFs, excluding the 17 nucleotides ploy (A) at the 3' end. Phylogenetic analysis based on part of RdRp gene of this strain showed that it was classified into subgroup GIII. Sequence alignment indicated that there was an inserted 21-nt long nucleotide sequence at the 3' end of ORF2. The insertion showed high antigenicity index comparing to other regions in ORF2.
Conclusion: Ch-sw-sav1 shared similar genetic profile with an American PEC strain except the 21-nt nucleotide at the 3' end of ORF2. The insert sequence shared high identity with part gene of Sus scrofa clone RP44-484M10.
Figures



Similar articles
-
Genomic organization and recombination analysis of a porcine sapovirus identified from a piglet with diarrhea in China.Virol J. 2017 Mar 16;14(1):57. doi: 10.1186/s12985-017-0729-1. Virol J. 2017. PMID: 28302145 Free PMC article.
-
Seroprevalence and molecular detection of porcine sapovirus in symptomatic suckling piglets in Guangdong Province, China.Trop Anim Health Prod. 2014 Mar;46(3):583-7. doi: 10.1007/s11250-013-0531-z. Epub 2014 Jan 10. Trop Anim Health Prod. 2014. PMID: 24407531
-
High genetic diversity in RdRp gene of Brazilian porcine sapovirus strains.Vet Microbiol. 2008 Sep 18;131(1-2):185-91. doi: 10.1016/j.vetmic.2008.02.021. Epub 2008 Mar 4. Vet Microbiol. 2008. PMID: 18403136
-
Porcine sapoviruses: Pathogenesis, epidemiology, genetic diversity, and diagnosis.Virus Res. 2020 Sep;286:198025. doi: 10.1016/j.virusres.2020.198025. Epub 2020 May 26. Virus Res. 2020. PMID: 32470356 Free PMC article. Review.
-
[Systematic review on the characteristics of acute gastroenteritis outbreaks caused by sapovirus].Zhonghua Liu Xing Bing Xue Za Zhi. 2019 Jan 10;40(1):93-98. doi: 10.3760/cma.j.issn.0254-6450.2019.01.019. Zhonghua Liu Xing Bing Xue Za Zhi. 2019. PMID: 30669739 Chinese.
Cited by
-
RT-PCR test for detecting porcine sapovirus in weanling piglets in Hunan Province, China.Trop Anim Health Prod. 2012 Oct;44(7):1335-9. doi: 10.1007/s11250-012-0138-9. Epub 2012 Apr 11. Trop Anim Health Prod. 2012. PMID: 22492394
-
High Genetic Diversity of Porcine Sapovirus From Diarrheic Piglets in Yunnan Province, China.Front Vet Sci. 2022 Jul 7;9:854905. doi: 10.3389/fvets.2022.854905. eCollection 2022. Front Vet Sci. 2022. PMID: 35873674 Free PMC article.
-
Serological and molecular investigation of porcine sapovirus infection in piglets in Xinjiang, China.Trop Anim Health Prod. 2016 Apr;48(4):863-9. doi: 10.1007/s11250-016-1023-8. Epub 2016 Feb 22. Trop Anim Health Prod. 2016. PMID: 26898687
-
Spatiotemporal trends in the discovery of new swine infectious agents.Vet Res. 2015 Sep 28;46:114. doi: 10.1186/s13567-015-0226-8. Vet Res. 2015. PMID: 26412219 Free PMC article. Review.
-
Complete genome of a porcine calicivirus strain in Anhui province, China, is significantly shorter than that of the other Chinese strain.J Virol. 2012 Dec;86(24):13823. doi: 10.1128/JVI.02613-12. J Virol. 2012. PMID: 23166238 Free PMC article.
References
-
- Bajolet O, Chippaux-Hyppolite C. Rotavirus and other viruses of diarrhea. Bull Soc Pathol Exot. 1998;91(5 Pt 1-2):432–437. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

