Crystal structure of CYP24A1, a mitochondrial cytochrome P450 involved in vitamin D metabolism
- PMID: 19961857
- PMCID: PMC2830900
- DOI: 10.1016/j.jmb.2009.11.057
Crystal structure of CYP24A1, a mitochondrial cytochrome P450 involved in vitamin D metabolism
Abstract
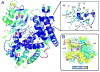
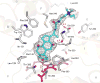
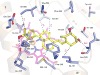
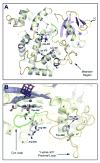
Cytochrome P450 (CYP) 24A1 catalyzes the side-chain oxidation of the hormonal form of vitamin D. Expression of CYP24A1 is up-regulated to attenuate vitamin D signaling associated with calcium homeostasis and cellular growth processes. The development of therapeutics for disorders linked to vitamin D insufficiency would be greatly facilitated by structural knowledge of CYP24A1. Here, we report the crystal structure of rat CYP24A1 at 2.5 A resolution. The structure exhibits an open cleft leading to the active-site heme prosthetic group on the distal surface that is likely to define the path of substrate access into the active site. The entrance to the cleft is flanked by conserved hydrophobic residues on helices A' and G', suggesting a mode of insertion into the inner mitochondrial membrane. A docking model for 1alpha,25-dihydroxyvitamin D(3) binding in the open form of CYP24A1 that clarifies the structural determinants of secosteroid recognition and validates the predictive power of existing homology models of CYP24A1 is proposed. Analysis of CYP24A1's proximal surface identifies the determinants of adrenodoxin recognition as a constellation of conserved residues from helices K, K'', and L that converge with an adjacent lysine-rich loop for binding the redox protein. Overall, the CYP24A1 structure provides the first template for understanding membrane insertion, substrate binding, and redox partner interaction in mitochondrial P450s.
2009 Elsevier Ltd. All rights reserved.
Figures

Similar articles
-
Hybrid homology modeling and mutational analysis of cytochrome P450C24A1 (CYP24A1) of the Vitamin D pathway: insights into substrate specificity and membrane bound structure-function.Arch Biochem Biophys. 2007 Apr 15;460(2):262-73. doi: 10.1016/j.abb.2006.11.018. Epub 2006 Dec 3. Arch Biochem Biophys. 2007. PMID: 17207766 Free PMC article.
-
Homology model of 1alpha,25-dihydroxyvitamin D3 24-hydroxylase cytochrome P450 24A1 (CYP24A1): active site architecture and ligand binding.J Steroid Biochem Mol Biol. 2007 Apr;104(1-2):53-60. doi: 10.1016/j.jsbmb.2006.09.041. Epub 2007 Jan 19. J Steroid Biochem Mol Biol. 2007. PMID: 17240137
-
Metabolism of vitamin D3 by cytochromes P450.Front Biosci. 2005 Jan 1;10:119-34. doi: 10.2741/1514. Print 2005 Jan 1. Front Biosci. 2005. PMID: 15574355 Review.
-
A new insight into the role of rat cytochrome P450 24A1 in metabolism of selective analogs of 1α,25-dihydroxyvitamin D₃.Arch Biochem Biophys. 2011 May 1;509(1):33-43. doi: 10.1016/j.abb.2011.02.004. Epub 2011 Feb 19. Arch Biochem Biophys. 2011. PMID: 21338573 Free PMC article.
-
25-Hydroxyvitamin D-24-hydroxylase (CYP24A1): its important role in the degradation of vitamin D.Arch Biochem Biophys. 2012 Jul 1;523(1):9-18. doi: 10.1016/j.abb.2011.11.003. Epub 2011 Nov 12. Arch Biochem Biophys. 2012. PMID: 22100522 Review.
Cited by
-
Rat CYP24A1 acts on 20-hydroxyvitamin D(3) producing hydroxylated products with increased biological activity.Biochem Pharmacol. 2012 Dec 15;84(12):1696-704. doi: 10.1016/j.bcp.2012.09.032. Epub 2012 Oct 5. Biochem Pharmacol. 2012. PMID: 23041230 Free PMC article.
-
Characterization of a Cleavable Fusion of Human CYP24A1 with Adrenodoxin Reveals the Variable Role of Hydrophobics in Redox Partner Binding.Biochemistry. 2022 Jan 18;61(2):57-66. doi: 10.1021/acs.biochem.1c00770. Epub 2022 Jan 3. Biochemistry. 2022. PMID: 34979083 Free PMC article.
-
Hydroxylation of 20-hydroxyvitamin D3 by human CYP3A4.J Steroid Biochem Mol Biol. 2016 May;159:131-41. doi: 10.1016/j.jsbmb.2016.03.014. Epub 2016 Mar 9. J Steroid Biochem Mol Biol. 2016. PMID: 26970587 Free PMC article.
-
Molecular dynamics, docking and quantum calculations reveal conformational changes influenced by CYP271A amino acid mutations related to cerebrotendinous xanthomatosis.Sci Rep. 2025 Mar 25;15(1):10229. doi: 10.1038/s41598-025-93966-7. Sci Rep. 2025. PMID: 40133480 Free PMC article.
-
A novel type of allosteric regulation: functional cooperativity in monomeric proteins.Arch Biochem Biophys. 2012 Mar 15;519(2):91-102. doi: 10.1016/j.abb.2011.12.017. Epub 2012 Jan 8. Arch Biochem Biophys. 2012. PMID: 22245335 Free PMC article. Review.
References
-
- Bernhardt R. Cytochromes P450 as versatile biocatalysts. J Biotechnol. 2006;124:128–145. Review. - PubMed
-
- Sigel A, Sigel H, Sigel R. In: Metal Ions in Life Sciences, Vol 3: The Ubiquitous Roles of Cytochrome P450 Proteins. Sigel A, Sigel H, Sigel R, editors. New Jersey: John Wiley and Sons, Ltd.; 2007.
-
- Nelson DR. Metazoan cytochrome P450 evolution. Comp Biochem Physiol C Pharmacol Toxicol Endocrinol. 1998;1-3:15–22. Review. - PubMed
-
- Guengerich FP. Mechanisms of cytochrome P450 substrate oxidation: MiniReview. J Biochem Mol Toxicol. 2007;21:163–168. Review. - PubMed
-
- Grinberg AV, Hannemann F, Schiffler B, Müller J, Heinemann U, Bernhardt R. Adrenodoxin: structure, stability, and electron transfer properties. Proteins. 2000;40:590–612. Review. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

