Novel Chlamydia pneumoniae vaccine candidates confirmed by Th1-enhanced genetic immunization
- PMID: 19961962
- PMCID: PMC2822074
- DOI: 10.1016/j.vaccine.2009.11.046
Novel Chlamydia pneumoniae vaccine candidates confirmed by Th1-enhanced genetic immunization
Abstract
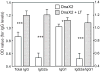
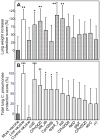
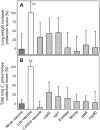
Identification of highly immunogenic antigens is critical for the construction of an efficacious subunit vaccine against Chlamydia pneumoniae infections. A previous project used a genome-wide screen to identify 12 protective C. pneumoniae candidate genes in an A/J mouse lung disease model (Li et al. [14]). Due to insufficient induction of Th1 immunity, these genes elicited only modest protection. Here, we used the Escherichia coli heat-labile enterotoxin as a Th1-enhancing genetic adjuvant, and re-tested these 12 genes, in parallel with six genes identified by other investigators. Vaccine candidate genes cutE and Cpn0420 conferred significant protection by all criteria evaluated (prevention of C. pneumoniae-induced death, reduction of lung disease, elimination of C. pneumoniae). Gene oppA_2 was protective by disease reduction and C. pneumoniae elimination. Four other genes were protective by a single criterion. None of the six genes reported elsewhere protected by reduction of lung disease or elimination of C. pneumoniae, but three protected by increasing survival.
Copyright (c) 2009 Elsevier Ltd. All rights reserved.
Figures
Similar articles
-
Mouse model of respiratory Chlamydia pneumoniae infection for a genomic screen of subunit vaccine candidates.Vaccine. 2006 Apr 5;24(15):2917-27. doi: 10.1016/j.vaccine.2005.12.035. Epub 2006 Jan 4. Vaccine. 2006. PMID: 16434129
-
Adjuvant modulation of the immune responses and the outcome of infection with Chlamydia pneumoniae.Clin Exp Immunol. 2002 Dec;130(3):393-403. doi: 10.1046/j.1365-2249.2002.02007.x. Clin Exp Immunol. 2002. PMID: 12452828 Free PMC article.
-
Adjuvant modulation of the immune response of mice against the LcrE protein of Chlamydophila pneumoniae.Int J Med Microbiol. 2009 Nov;299(7):520-8. doi: 10.1016/j.ijmm.2009.04.002. Epub 2009 May 17. Int J Med Microbiol. 2009. PMID: 19451031
-
The adjuvant double mutant Escherichia coli heat labile toxin enhances IL-17A production in human T cells specific for bacterial vaccine antigens.PLoS One. 2012;7(12):e51718. doi: 10.1371/journal.pone.0051718. Epub 2012 Dec 20. PLoS One. 2012. PMID: 23284753 Free PMC article.
-
Contemporary approaches to designing and evaluating vaccines against Chlamydia.Expert Rev Vaccines. 2003 Feb;2(1):129-46. doi: 10.1586/14760584.2.1.129. Expert Rev Vaccines. 2003. PMID: 12901604 Review.
Cited by
-
Seventy Years of Chlamydia Vaccine Research - Limitations of the Past and Directions for the Future.Front Microbiol. 2019 Jan 31;10:70. doi: 10.3389/fmicb.2019.00070. eCollection 2019. Front Microbiol. 2019. PMID: 30766521 Free PMC article. Review.
References
-
- Sessa R, Nicoletti M, Di Pietro M, Schiavoni G, Santino I, Zagaglia G, et al. Chlamydia pneumoniae and atherosclerosis: current state and future prospectives. Int J Immunopathol Pharmacol. 2009;22:9–14. - PubMed
-
- Saikku P. Chlamydia pneumoniae in atherosclerosis. J Intern Med. 2000;247:391–396. - PubMed
-
- de Kruif MD, van Gorp EC, Keller TT, Ossewaarde JM, Cate HT. Chlamydia pneumoniae infections in mouse models: relevance for atherosclerosis research. Cardiovasc Res. 2005;65:317–27. - PubMed
-
- Longbottom D. Chlamydial vaccine development. J Med Microbiol. 2003;52:537–40. - PubMed
-
- Wang C, Gao D, Kaltenboeck B. Acute Chlamydia pneumoniae reinfection accelerates the development of insulin resistance and diabetes in obese C57BL/6 mice. J Infect Dis. 2009;200:279–87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

