Differential transcriptomic responses of Biomphalaria glabrata (Gastropoda, Mollusca) to bacteria and metazoan parasites, Schistosoma mansoni and Echinostoma paraensei (Digenea, Platyhelminthes)
- PMID: 19962194
- PMCID: PMC2814977
- DOI: 10.1016/j.molimm.2009.10.019
Differential transcriptomic responses of Biomphalaria glabrata (Gastropoda, Mollusca) to bacteria and metazoan parasites, Schistosoma mansoni and Echinostoma paraensei (Digenea, Platyhelminthes)
Abstract
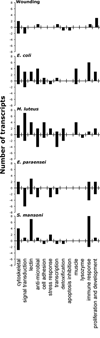
A 70-mer-oligonucleotide-based microarray (1152 features) that emphasizes stress and immune responses factors was constructed to study transcriptomic responses of the snail Biomphalaria glabrata to different immune challenges. In addition to sequences with relevant putative ID and Gene Ontology (GO) annotation, the array features non-immune factors and unknown B. glabrata ESTs for functional gene discovery. The transcription profiles of B. glabrata (3 biological replicates, each a pool of 5 snails) were recorded at 12h post-wounding, exposure to Gram negative or Gram positive bacteria (Escherichia coli and Micrococcus luteus, respectively), or infection with compatible trematode parasites (Schistosoma mansoni or Echinostoma paraensei, 20 miracidia/snail), relative to controls, using universal reference RNA. The data were subjected to Significance Analysis for Microarrays (SAM), with a false positive rate (FPR) <or=10%. Wounding yielded a modest differential expression profile (27 up/21 down) with affected features mostly dissimilar from other treatments. Partially overlapping, yet distinct expression profiles were recorded from snails challenged with E. coli (83 up/20 down) or M. luteus (120 up/42 down), mostly showing up-regulation of defense and stress-related features. Significantly altered expression of selected immune features indicates that B. glabrata detects and responds differently to compatible trematodes. Echinostoma paraensei infection was associated mostly with down-regulation of many (immune-) transcripts (42 up/68 down), whereas S. mansoni exposure yielded a preponderance of up-regulated features (140 up/23 down), with only few known immune genes affected. These observations may reflect the divergent strategies developed by trematodes during their evolution as specialized pathogens of snails to negate host defense responses. Clearly, the immune defenses of B. glabrata distinguish and respond differently to various immune challenges.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures





References
-
- Abraham EG, Islam S, Srinivasan P, Ghosh AK, Valenzuela JG, Ribeiro JM, Kafatos FC, Dimopoulos G, Jacobs-Lorena M. Analysis of the Plasmodium and Anopheles transcriptional repertoire during ookinete development and midgut invasion. J Biol Chem. 2004;279:5573–5580. - PubMed
-
- Archambault V, Glover DM. Polo-like kinases: conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol. 2009;10:265–275. - PubMed
-
- Azumi K, De Santis R, De Tomaso A, Rigoutsos I, Yoshizaki F, Pinto MR, Marino R, Shida K, Ikeda M, Arai M, Inoue Y, Shimizu T, Satoh N, Rokhsar DS, Du Pasquier L, Kasahara M, Satake M, Nonaka M. Genomic analysis of immunity in a Urochordate and the emergence of the vertebrate immune system: "waiting for Godot". Immunogenetics. 2003;55:570–581. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

