Structural and biophysical studies of human PARP-1 in complex with damaged DNA
- PMID: 19962992
- PMCID: PMC2864582
- DOI: 10.1016/j.jmb.2009.11.062
Structural and biophysical studies of human PARP-1 in complex with damaged DNA
Abstract
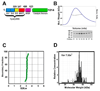
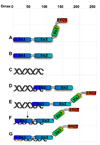
The enzyme poly(ADP-ribose) polymerase-1 (PARP-1) is a global monitor of chromatin structure and DNA damage repair. PARP-1 binds to nucleosomes and poly(ADP-ribosylates) histones and several chromatin-associated factors to expose specific DNA sequences to the cellular machinery involved in gene transcription and/or DNA damage repair. While these processes are critical to genomic stability, the molecular mechanisms of how DNA damage induces PARP-1 activation are poorly understood. We have used biochemical and thermodynamic measurements in conjunction with small-angle X-ray scattering to determine the stoichiometry, affinity, and overall structure of a human PARP-1 construct containing the entire DNA binding region, the zinc ribbon domain, and automodification domains (residues 1-486). The interaction of this PARP-1 protein construct with three different DNA damage models (DNA constructs containing a nick, a blunt end, or a 3' extension) was evaluated. Our data indicate that PARP-1 binds each DNA damage model as a monomer and with similar affinity, in all cases resulting in robust activation of the catalytic domain. Using small-angle X-ray scattering, we determined that the N-terminal half of PARP-1 behaves as an extended and flexible arrangement of individually folded domains in the absence of DNA. Upon binding DNA, PARP-1 undergoes a conformational change in the area surrounding the zinc ribbon domain. These data support a model in which PARP-1, upon binding DNA, undergoes a conformational change to become an active nuclear enzyme.
Copyright 2009 Elsevier Ltd. All rights reserved.
Figures




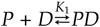
References
-
- Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. - PubMed
-
- Bouchard VJ, Rouleau M, Poirier GG. PARP-1, a determinant of cell survival in response to DNA damage. Exp Hematol. 2003;31:446–454. - PubMed
-
- Jacobson EL, Antol KM, Juarez-Salinas H, Jacobson MK. Poly(ADP-ribose) metabolism in ultraviolet irradiated human fibroblasts. J Biol Chem. 1983;258:103–107. - PubMed
-
- Oleinick NL, Evans HH. Poly(ADP-ribose) and the response of cells to ionizing radiation. Radiat Res. 1985;101:29–46. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

