DeNAno: Selectable deoxyribonucleic acid nanoparticle libraries
- PMID: 19963022
- PMCID: PMC2822443
- DOI: 10.1016/j.jbiotec.2009.12.002
DeNAno: Selectable deoxyribonucleic acid nanoparticle libraries
Abstract
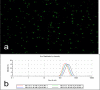
DNA nanoparticles of approximately 250 nm were produced by rolling circle replication of circular oligonucleotide templates which results in highly condensed DNA particulates presenting concatemeric sequence repeats. Using templates containing randomized sequences, high diversity libraries of particles were produced. A biopanning method that iteratively screens for binding and uses PCR to recover selected particles was developed. The initial application of this technique was the selection of particles that bound to human dendritic cells (DCs). Following nine rounds of selection the population of particles was enriched for particles that bound DCs, and individual binding clones were isolated and confirmed by flow cytometry and microscopy. This process, which we have termed DeNAno, represents a novel library technology akin to aptamer and phage display, but unique in that the selected moiety is a multivalent nanoparticle whose activity is intrinsic to its sequence. Cell targeted DNA nanoparticles may have applications in cell imaging, cell sorting, and cancer therapy.
Copyright 2009 Elsevier B.V. All rights reserved.
Figures
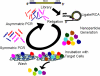


References
-
- Ferrari M. Cancer nanotechnology: opportunities and challenges. Nat Rev Cancer. 2005;5:161–171. - PubMed
-
- Wilson DS, Szostak JW. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999;68:611–647. - PubMed
-
- Smith GP, Scott JK. Libraries of peptides and proteins displayed on filamentous phage. Methods Enzymol. 1993;217:228–257. - PubMed
-
- Clackson T, Hoogenboom HR, Griffiths AD, Winter G. Making antibody fragments using phage display libraries. Nature. 1991;352:624–628. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

