Mouse hepatic lipase alleles with variable effects on lipoprotein composition and size
- PMID: 19965617
- PMCID: PMC2853430
- DOI: 10.1194/jlr.M002378
Mouse hepatic lipase alleles with variable effects on lipoprotein composition and size
Abstract
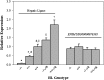
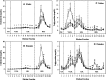
The structural features responsible for the activities of hepatic lipase (HL) can be clarified by in vivo comparisons of naturally occurring variants. The coding sequence of HL from C57BL/6J (B6) and SPRET/EiJ (SPRET) mice differs by four amino acids (S106N, A156V, L416V, S480T); however, these changes are not predicted to influence HL function. To test for allelic effects, we generated SPRET-HL transgenics with physiological levels of HL mRNA and HL activity that was parallel in female transgenics and about 70% higher in male transgenics, toward tri-[3H]oleate, compared with B6 controls. We found no correlation between activity levels and plasma lipids. However, significant allelic effects on plasma lipids were observed. Compared with B6-HL, SPRET-HL mediated reductions in total cholesterol (TC) and VLDL-, LDL- and HDL-cholesterol and HDL-triglyceride (TG) in fed males, and SPRET-HL decreased total TG and VLDL- and HDL-TG levels in fasted males. Fasted female transgenics had reduced TC compared with controls. We also found allele and sex effects on lipoprotein particle size. Male transgenic mice had increased VLDL and decreased LDL size, and female transgenic mice had decreased HDL size compared with control animals. These findings demonstrate highly divergent effects of naturally occurring HL coding sequence variants on lipid and lipoprotein metabolism.
Figures


References
-
- Busch S. J., Barnhart R. L., Martin G. A., Fitzgerald M. C., Yates M. T., Mao S. J., Thomas C. E., Jackson R. L. 1994. Human hepatic triglyceride lipase expression reduces high density lipoprotein and aortic cholesterol in cholesterol-fed transgenic mice. J. Biol. Chem. 269: 16376–16382. - PubMed
-
- Homanics G. E., de Silva H. V., Osada J., Zhang S. H., Wong H., Borensztajn J., Maeda N. 1995. Mild dyslipidemia in mice following targeted inactivation of the hepatic lipase gene. J. Biol. Chem. 270: 2974–2980. - PubMed
-
- Jensen G. L., Daggy B., Bensadoun A. 1982. Triacylglycerol lipase, monoacylglycerol lipase and phospholipase activities of highly purified rat hepatic lipase. Biochim. Biophys. Acta. 710: 464–470. - PubMed
-
- Little J. A., Connelly P. W. 1986. Familial hepatic lipase deficiency. Adv. Exp. Med. Biol. 201: 253–260. - PubMed
-
- Dichek H. L., Qian K., Agrawal N. 2004. The bridging function of hepatic lipase clears plasma cholesterol in LDL receptor-deficient “apoB-48-only” and “apoB-100-only” mice. J. Lipid Res. 45: 551–560. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

