Comparison of gene expression profiles between human and mouse monocyte subsets
- PMID: 19965649
- PMCID: PMC2810986
- DOI: 10.1182/blood-2009-07-235028
Comparison of gene expression profiles between human and mouse monocyte subsets
Erratum in
- Blood. 2010 Aug 5;116(5):857
Abstract
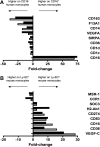
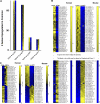
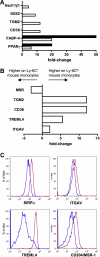
Blood of both humans and mice contains 2 main monocyte subsets. Here, we investigated the extent of their similarity using a microarray approach. Approximately 270 genes in humans and 550 genes in mice were differentially expressed between subsets by 2-fold or more. More than 130 of these gene expression differences were conserved between mouse and human monocyte subsets. We confirmed numerous of these differences at the cell surface protein level. Despite overall conservation, some molecules were conversely expressed between the 2 species' subsets, including CD36, CD9, and TREM-1. Other differences included a prominent peroxisome proliferator-activated receptor gamma (PPARgamma) signature in mouse monocytes, which is absent in humans, and strikingly opposed patterns of receptors involved in uptake of apoptotic cells and other phagocytic cargo between human and mouse monocyte subsets. Thus, whereas human and mouse monocyte subsets are far more broadly conserved than currently recognized, important differences between the species deserve consideration when models of human disease are studied in mice.
Figures




References
-
- Passlick B, Flieger D, Ziegler-Heitbrock HW. Identification and characterization of a novel monocyte subpopulation in human peripheral blood. Blood. 1989;74(7):2527–2534. - PubMed
-
- Ziegler-Heitbrock L. The CD14+ CD16+ blood monocytes: their role in infection and inflammation. J Leukoc Biol. 2007;81(3):584–592. - PubMed
-
- Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 2003;19(1):71–82. - PubMed
-
- Sunderkötter C, Nikolic T, Dillon MJ, et al. Subpopulations of mouse blood monocytes differ in maturation stage and inflammatory response. J Immunol. 2004;172(7):4410–4417. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

