Regulation of human NK-cell cytokine and chemokine production by target cell recognition
- PMID: 19965656
- PMCID: PMC2844017
- DOI: 10.1182/blood-2009-08-238469
Regulation of human NK-cell cytokine and chemokine production by target cell recognition
Abstract
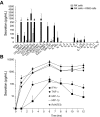
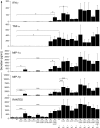
Natural killer (NK)-cell recognition of infected or neoplastic cells can induce cytotoxicity and cytokine secretion. So far, it has been difficult to assess the relative contribution of multiple NK-cell activation receptors to cytokine and chemokine production upon target cell recognition. Using Drosophila cells expressing ligands for the NK-cell receptors LFA-1, NKG2D, DNAM-1, 2B4, and CD16, we studied the minimal requirements for secretion by freshly isolated, human NK cells. Target cell stimulation induced secretion of predominately proinflammatory cytokines and chemokines. Release of chemokines MIP-1alpha, MIP-1beta, and RANTES was induced within 1 hour of stimulation, whereas release of TNF-alpha and IFN-gamma occurred later. Engagement of CD16, 2B4, or NKG2D sufficed for chemokine release, whereas induction of TNF-alpha and IFN-gamma required engagement of additional receptors. Remarkably, our results revealed that, upon target cell recognition, CD56(dim) NK cells were more prominent cytokine and chemokine producers than CD56(bright) NK cells. The present data demonstrate how specific target cell ligands dictate qualitative and temporal aspects of NK-cell cytokine and chemokine responses. Conceptually, the results point to CD56(dim) NK cells as an important source of cytokines and chemokines upon recognition of aberrant cells, producing graded responses depending on the multiplicity of activating receptors engaged.
Figures

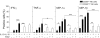
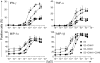

Comment in
-
Target recognition-induced NK-cell responses.Blood. 2010 Mar 18;115(11):2119-20. doi: 10.1182/blood-2010-01-260448. Blood. 2010. PMID: 20299516 No abstract available.
References
-
- Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. - PubMed
-
- Bottino C, Castriconi R, Moretta L, Moretta A. Cellular ligands of activating NK receptors. Trends Immunol. 2005;26(4):221–226. - PubMed
-
- Lee SH, Miyagi T, Biron CA. Keeping NK cells in highly regulated antiviral warfare. Trends Immunol. 2007;28(6):252–259. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

