Optimization of minimum set of protein-DNA interactions: a quasi exact solution with minimum over-fitting
- PMID: 19965883
- PMCID: PMC2815656
- DOI: 10.1093/bioinformatics/btp664
Optimization of minimum set of protein-DNA interactions: a quasi exact solution with minimum over-fitting
Abstract
Motivation: A major limitation in modeling protein interactions is the difficulty of assessing the over-fitting of the training set. Recently, an experimentally based approach that integrates crystallographic information of C2H2 zinc finger-DNA complexes with binding data from 11 mutants, 7 from EGR finger I, was used to define an improved interaction code (no optimization). Here, we present a novel mixed integer programming (MIP)-based method that transforms this type of data into an optimized code, demonstrating both the advantages of the mathematical formulation to minimize over- and under-fitting and the robustness of the underlying physical parameters mapped by the code.
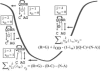
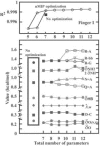
Results: Based on the structural models of feasible interaction networks for 35 mutants of EGR-DNA complexes, the MIP method minimizes the cumulative binding energy over all complexes for a general set of fundamental protein-DNA interactions. To guard against over-fitting, we use the scalability of the method to probe against the elimination of related interactions. From an initial set of 12 parameters (six hydrogen bonds, five desolvation penalties and a water factor), we proceed to eliminate five of them with only a marginal reduction of the correlation coefficient to 0.9983. Further reduction of parameters negatively impacts the performance of the code (under-fitting). Besides accurately predicting the change in binding affinity of validation sets, the code identifies possible context-dependent effects in the definition of the interaction networks. Yet, the approach of constraining predictions to within a pre-selected set of interactions limits the impact of these potential errors to related low-affinity complexes.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures
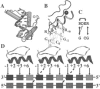



References
-
- Bae KH, et al. Human zinc fingers as building blocks in the construction of artificial transcription factors. Nat. Biotechnol. 2003;21:275–280. - PubMed
-
- Bonvin AM, et al. Water molecules in DNA recognition II: a molecular dynamics view of the structure and hydration of the trp operator. J. Mol. Biol. 1998;282:859–873. - PubMed
-
- Bueno M, Camacho CJ. Acidic groups docked to well defined wetted pockets at the core of the binding interface: a tale of scoring and missing protein interactions in CAPRI. Proteins. 2007a;69:786–792. - PubMed
-
- Bueno M, et al. SIMPLE estimate of the free energy change due to aliphatic mutations: superior predictions based on first principles. Proteins. 2007b;68:850–862. - PubMed
-
- Camacho CJ, et al. Scoring a diverse set of high-quality docked conformations: a metascore based on electrostatic and desolvation interactions. Proteins. 2006;63:868–877. - PubMed

