Functional characterization of poplar wood-associated NAC domain transcription factors
- PMID: 19965968
- PMCID: PMC2815876
- DOI: 10.1104/pp.109.148270
Functional characterization of poplar wood-associated NAC domain transcription factors
Abstract
Wood is the most abundant biomass produced by land plants. Dissection of the molecular mechanisms underlying the transcriptional regulation of wood formation is a fundamental issue in plant biology and has important implications in tree biotechnology. Although a number of transcription factors in tree species have been shown to be associated with wood formation and some of them are implicated in lignin biosynthesis, none of them have been demonstrated to be key regulators of the biosynthesis of all three major components of wood. In this report, we have identified a group of NAC domain transcription factors, PtrWNDs, that are preferentially expressed in developing wood of poplar (Populus trichocarpa). Expression of PtrWNDs in the Arabidopsis (Arabidopsis thaliana) snd1 nst1 double mutant effectively complemented the secondary wall defects in fibers, indicating that PtrWNDs are capable of activating the entire secondary wall biosynthetic program. Overexpression of PtrWND2B and PtrWND6B in Arabidopsis induced the expression of secondary wall-associated transcription factors and secondary wall biosynthetic genes and, concomitantly, the ectopic deposition of cellulose, xylan, and lignin. Furthermore, PtrWND2B and PtrWND6B were able to activate the promoter activities of a number of poplar wood-associated transcription factors and wood biosynthetic genes. Together, these results demonstrate that PtrWNDs are functional orthologs of SND1 and suggest that PtrWNDs together with their downstream transcription factors form a transcriptional network involved in the regulation of wood formation in poplar.
Figures


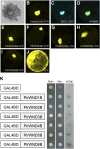
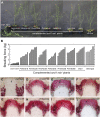


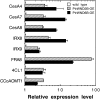
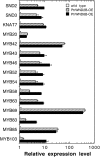


Comment in
-
The poplar PtrWNDs are transcriptional activators of secondary cell wall biosynthesis.Plant Signal Behav. 2010 Apr;5(4):469-72. doi: 10.4161/psb.5.4.11400. Epub 2010 Apr 2. Plant Signal Behav. 2010. PMID: 20383071 Free PMC article.
References
-
- Andersson-Gunneras S, Mellerowicz EJ, Love J, Segerman B, Ohmiya Y, Coutinho PM, Nilsson P, Henrissat B, Moritz T, Sundberg B (2006) Biosynthesis of cellulose-enriched tension wood in Populus: global analysis of transcripts and metabolites identifies biochemical and developmental regulators in secondary wall biosynthesis. Plant J 45 144–165 - PubMed
-
- Bechtold N, Bouchez D (1994) In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. In I Potrykus, G Spangenberg, eds, Gene Transfer to Plants. Springer-Verlag, Berlin, pp 19–23 - PubMed
-
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54 519–546 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

