Opposing regulatory roles of phosphorylation and acetylation in DNA mispair processing by thymine DNA glycosylase
- PMID: 19966277
- PMCID: PMC2831317
- DOI: 10.1093/nar/gkp1097
Opposing regulatory roles of phosphorylation and acetylation in DNA mispair processing by thymine DNA glycosylase
Abstract
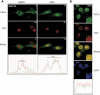
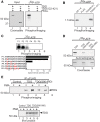
CpG dinucleotides are mutational hotspots associated with cancer and genetic diseases. Thymine DNA glycosylase (TDG) plays an integral role in CpG maintenance by excising mispaired thymine and uracil in a CpG context and also participates in transcriptional regulation via gene-specific CpG demethylation and functional interactions with the transcription machinery. Here, we report that protein kinase C alpha (PKCalpha) interacts with TDG and phosphorylates amino-terminal serine residues adjacent to lysines acetylated by CREB-binding protein (CBP) and p300 (CBP/p300). We establish that acetylation and phosphorylation are mutually exclusive, and their interplay dramatically alters the DNA mispair-processing functions of TDG. Remarkably, acetylation of the amino-terminal region abrogates high-affinity DNA binding and selectively prevents processing of G:T mispairs. In contrast, phosphorylation does not markedly alter DNA interactions, but may preserve G:T processing in vivo by preventing CBP-mediated acetylation. Mutational analysis suggests that the acetyl-acceptor lysines are not directly involved in contacting DNA, but may constitute a conformationally sensitive interface that modulates DNA interactions. These findings reveal opposing roles of CBP/p300 and PKCalpha in regulating the DNA repair functions of TDG and suggest that the interplay of these modifications in vivo may be critically important in the maintenance of CpG dinucleotides and epigenetic regulation.
Figures





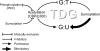
Similar articles
-
Histone deacetylase SIRT1 modulates and deacetylates DNA base excision repair enzyme thymine DNA glycosylase.Biochem J. 2013 Nov 15;456(1):89-98. doi: 10.1042/BJ20130670. Biochem J. 2013. PMID: 23952905 Free PMC article.
-
Role of base excision repair in maintaining the genetic and epigenetic integrity of CpG sites.DNA Repair (Amst). 2015 Aug;32:33-42. doi: 10.1016/j.dnarep.2015.04.011. Epub 2015 May 1. DNA Repair (Amst). 2015. PMID: 26021671 Free PMC article. Review.
-
SUMO-1-dependent allosteric regulation of thymine DNA glycosylase alters subnuclear localization and CBP/p300 recruitment.Mol Cell Biol. 2007 Jan;27(1):229-43. doi: 10.1128/MCB.00323-06. Epub 2006 Oct 23. Mol Cell Biol. 2007. PMID: 17060459 Free PMC article.
-
Interaction with the DNA Repair Protein Thymine DNA Glycosylase Regulates Histone Acetylation by p300.Biochemistry. 2016 Dec 13;55(49):6766-6775. doi: 10.1021/acs.biochem.6b00841. Epub 2016 Dec 1. Biochemistry. 2016. PMID: 27951654 Free PMC article.
-
The enigmatic thymine DNA glycosylase.DNA Repair (Amst). 2007 Apr 1;6(4):489-504. doi: 10.1016/j.dnarep.2006.10.013. Epub 2006 Nov 20. DNA Repair (Amst). 2007. PMID: 17116428 Review.
Cited by
-
Acetylation of human TCF4 (TCF7L2) proteins attenuates inhibition by the HBP1 repressor and induces a conformational change in the TCF4::DNA complex.PLoS One. 2013 Apr 15;8(4):e61867. doi: 10.1371/journal.pone.0061867. Print 2013. PLoS One. 2013. PMID: 23613959 Free PMC article.
-
Repair of oxidative DNA damage and cancer: recent progress in DNA base excision repair.Antioxid Redox Signal. 2014 Feb 1;20(4):708-26. doi: 10.1089/ars.2013.5529. Epub 2013 Oct 15. Antioxid Redox Signal. 2014. PMID: 23901781 Free PMC article. Review.
-
Nucleosomes and the three glycosylases: High, medium, and low levels of excision by the uracil DNA glycosylase superfamily.DNA Repair (Amst). 2018 Dec;72:56-63. doi: 10.1016/j.dnarep.2018.09.008. Epub 2018 Sep 20. DNA Repair (Amst). 2018. PMID: 30268365 Free PMC article.
-
Histone deacetylase SIRT1 modulates and deacetylates DNA base excision repair enzyme thymine DNA glycosylase.Biochem J. 2013 Nov 15;456(1):89-98. doi: 10.1042/BJ20130670. Biochem J. 2013. PMID: 23952905 Free PMC article.
-
Role of base excision repair in maintaining the genetic and epigenetic integrity of CpG sites.DNA Repair (Amst). 2015 Aug;32:33-42. doi: 10.1016/j.dnarep.2015.04.011. Epub 2015 May 1. DNA Repair (Amst). 2015. PMID: 26021671 Free PMC article. Review.
References
-
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 2006;31:89–97. - PubMed
-
- Pfeifer GP. Mutagenesis at methylated CpG sequences. Curr. Top Microbiol. Immunol. 2006;301:259–281. - PubMed
-
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. - PubMed
-
- Pfeifer GP. p53 mutational spectra and the role of methylated CpG sequences. Mutat. Res. 2000;450:155–166. - PubMed
-
- Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum. Mutat. 2007;28:622–629. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

