Impairment of transforming growth factor beta signaling in caveolin-1-deficient hepatocytes: role in liver regeneration
- PMID: 19966340
- PMCID: PMC2823504
- DOI: 10.1074/jbc.M109.072900
Impairment of transforming growth factor beta signaling in caveolin-1-deficient hepatocytes: role in liver regeneration
Abstract
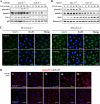
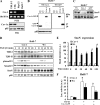
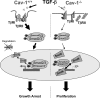
Caveolin-1 (Cav-1) is the main structural protein of caveolae and plays an important role in various cellular processes such as vesicular transport, cholesterol homeostasis, and signal transduction pathways. The expression and functional role of Cav-1 have been reported in liver and in hepatocyte cell lines, in human cirrhotic liver, and in hepatocellular carcinomas. Previous studies demonstrated that Cav-1 was dispensable for liver regeneration, because Cav-1(-/-) animals survived and fully regenerated liver function and size after partial hepatectomy. In this study, we have investigated the mechanisms by which the lack of Cav-1 accelerates liver regeneration after partial hepatectomy. The data show that transforming growth factor beta (TGF-beta) signaling is impaired in regenerating liver of Cav-1(-/-) mice and in hepatocytes derived from these animals. TGF-beta receptors I and II do not colocalize in the same membrane fraction in the hepatocytes derived from Cav-1(-/-) mice, as Smad2/3 signaling decreased in the absence of Cav-1 at the time that the transcriptional corepressor SnoN accumulates. Accordingly, the expression of TGF-beta target genes, such as plasminogen activator inhibitor-1, is decreased due to the presence of the high levels of SnoN. Moreover, hepatocyte growth factor inhibited TGF-beta signaling in the absence of Cav-1 by increasing SnoN expression. Taken together, these data might help to unravel why Cav-1-deficient mice exhibit an accelerated liver regeneration after partial hepatectomy and add new insights on the molecular mechanisms controlling the initial commitment to hepatocyte proliferation.
Figures




Similar articles
-
Caveolin-2 is a negative regulator of anti-proliferative function and signaling of transforming growth factor-β in endothelial cells.Am J Physiol Cell Physiol. 2011 Nov;301(5):C1161-74. doi: 10.1152/ajpcell.00486.2010. Epub 2011 Aug 10. Am J Physiol Cell Physiol. 2011. PMID: 21832243 Free PMC article.
-
Dispensability and dynamics of caveolin-1 during liver regeneration and in isolated hepatic cells.Hepatology. 2007 Sep;46(3):813-22. doi: 10.1002/hep.21746. Hepatology. 2007. PMID: 17654701
-
Up-regulated transcriptional repressors SnoN and Ski bind Smad proteins to antagonize transforming growth factor-beta signals during liver regeneration.J Biol Chem. 2002 Aug 9;277(32):28483-90. doi: 10.1074/jbc.M202403200. Epub 2002 May 21. J Biol Chem. 2002. PMID: 12023281
-
Caveolin-1, transforming growth factor-beta receptor internalization, and the pathogenesis of systemic sclerosis.Curr Opin Rheumatol. 2008 Nov;20(6):713-9. doi: 10.1097/bor.0b013e3283103d27. Curr Opin Rheumatol. 2008. PMID: 18949888 Free PMC article. Review.
-
Caveolin-1: a critical regulator of lung injury.Am J Physiol Lung Cell Mol Physiol. 2011 Feb;300(2):L151-60. doi: 10.1152/ajplung.00170.2010. Epub 2010 Nov 19. Am J Physiol Lung Cell Mol Physiol. 2011. PMID: 21097526 Free PMC article. Review.
Cited by
-
SnoN in regulation of embryonic development and tissue morphogenesis.FEBS Lett. 2012 Jul 4;586(14):1971-6. doi: 10.1016/j.febslet.2012.03.005. Epub 2012 Mar 10. FEBS Lett. 2012. PMID: 22710172 Free PMC article. Review.
-
Protein Tyrosine Phosphatase 1B (PTP1B) deficiency accelerates hepatic regeneration in mice.Am J Pathol. 2011 Apr;178(4):1591-604. doi: 10.1016/j.ajpath.2010.12.020. Epub 2011 Mar 4. Am J Pathol. 2011. PMID: 21406170 Free PMC article.
-
The less-often-traveled surface of stem cells: caveolin-1 and caveolae in stem cells, tissue repair and regeneration.Stem Cell Res Ther. 2013 Jul 30;4(4):90. doi: 10.1186/scrt276. Stem Cell Res Ther. 2013. PMID: 23899671 Free PMC article. Review.
-
Redox-induced Src kinase and caveolin-1 signaling in TGF-β1-initiated SMAD2/3 activation and PAI-1 expression.PLoS One. 2011;6(7):e22896. doi: 10.1371/journal.pone.0022896. Epub 2011 Jul 28. PLoS One. 2011. PMID: 21829547 Free PMC article.
-
Adipocyte SIRT1 knockout promotes PPARγ activity, adipogenesis and insulin sensitivity in chronic-HFD and obesity.Mol Metab. 2015 Mar 5;4(5):378-91. doi: 10.1016/j.molmet.2015.02.007. eCollection 2015 May. Mol Metab. 2015. PMID: 25973386 Free PMC article.
References
-
- Razani B., Woodman S. E., Lisanti M. P. (2002) Pharmacol. Rev. 54, 431–467 - PubMed
-
- Schnitzer J. E. (2001) Adv. Drug Delivery Rev. 49, 265–280 - PubMed
-
- Fu Y., Hoang A., Escher G., Parton R. G., Krozowski Z., Sviridov D. (2004) J. Biol. Chem. 279, 14140–14146 - PubMed
-
- Okamoto T., Schlegel A., Scherer P. E., Lisanti M. P. (1998) J. Biol. Chem. 273, 5419–5422 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

