Stretching chromatin through confinement
- PMID: 19967112
- PMCID: PMC3746321
- DOI: 10.1039/b909217j
Stretching chromatin through confinement
Abstract
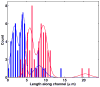
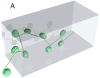
We present a method for the stretching of chromatin molecules in nanofluidic channels width a cross-section of about 80 x 80 nm(2), and hundreds of microns long. The stretching of chromatin to about 12 basepairs/nm enables location-resolved optical investigation of the nucleic material with a resolution of up to 6 kbp. The stretching is based on the equilibrium elongation that polymers experience when they are introduced into nanofluidic channels that are narrower than the Flory coil corresponding to the whole chromatin molecule. We investigate whether the elongation of reconstituted chromatin can be described by the de Gennes model. We compare nanofluidic stretching of bare DNA and chromatin of equal genomic length, and find that chromatin is 2.5 times more compact in its stretched state.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

