Farnesylcysteine lyase is involved in negative regulation of abscisic acid signaling in Arabidopsis
- PMID: 19969520
- PMCID: PMC2807925
- DOI: 10.1093/mp/ssp091
Farnesylcysteine lyase is involved in negative regulation of abscisic acid signaling in Arabidopsis
Abstract
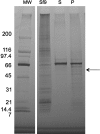
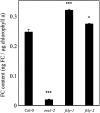
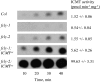
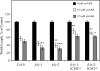
The Arabidopsis FCLY gene encodes a specific farnesylcysteine (FC) lyase, which is responsible for the oxidative metabolism of FC to farnesal and cysteine. In addition, fcly mutants with quantitative decreases in FC lyase activity exhibit an enhanced response to ABA. However, the enzymological properties of the FCLY-encoded enzyme and its precise role in ABA signaling remain unclear. Here, we show that recombinant Arabidopsis FC lyase expressed in insect cells exhibits high selectivity for FC as a substrate and requires FAD and molecular oxygen for activity. Arabidopsis FC lyase is also shown to undergo post-translational N-glycosylation. FC, which is a competitive inhibitor of isoprenylcysteine methyltransferase (ICMT), accumulates in fcly mutants. Moreover, the enhanced response of fcly mutants to ABA is reversed by ICMT overexpression. These observations support the hypothesis that the ABA hypersensitive phenotype of fcly plants is the result of FC accumulation and inhibition of ICMT.
Figures

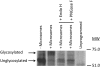


Similar articles
-
Arabidopsis thaliana plants possess a specific farnesylcysteine lyase that is involved in detoxification and recycling of farnesylcysteine.Plant J. 2007 Jun;50(5):839-47. doi: 10.1111/j.1365-313X.2007.03091.x. Epub 2007 Apr 8. Plant J. 2007. PMID: 17425716
-
Isoprenylcysteine methylation and demethylation regulate abscisic acid signaling in Arabidopsis.Plant Cell. 2008 Oct;20(10):2714-28. doi: 10.1105/tpc.107.053389. Epub 2008 Oct 28. Plant Cell. 2008. PMID: 18957507 Free PMC article.
-
The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA.Plant Physiol. 2006 Jan;140(1):127-39. doi: 10.1104/pp.105.070318. Epub 2005 Dec 16. Plant Physiol. 2006. PMID: 16361522 Free PMC article.
-
Arabidopsis zinc-finger protein 2 is a negative regulator of ABA signaling during seed germination.J Plant Physiol. 2010 Nov 1;167(16):1418-21. doi: 10.1016/j.jplph.2010.05.010. Epub 2010 Jul 8. J Plant Physiol. 2010. PMID: 20619483
-
Arabidopsis cysteine-rich receptor-like kinase 45 functions in the responses to abscisic acid and abiotic stresses.Plant Physiol Biochem. 2013 Jun;67:189-98. doi: 10.1016/j.plaphy.2013.03.013. Epub 2013 Mar 26. Plant Physiol Biochem. 2013. PMID: 23583936
Cited by
-
A Combination of Histological, Physiological, and Proteomic Approaches Shed Light on Seed Desiccation Tolerance of the Basal Angiosperm Amborella trichopoda.Proteomes. 2017 Jul 28;5(3):19. doi: 10.3390/proteomes5030019. Proteomes. 2017. PMID: 28788068 Free PMC article.
-
Genetic Dissection of Root Angle of Brassica napus in Response to Low Phosphorus.Front Plant Sci. 2021 Jul 29;12:697872. doi: 10.3389/fpls.2021.697872. eCollection 2021. Front Plant Sci. 2021. PMID: 34394150 Free PMC article.
-
Transcriptomes Divergence of Ricotia lunaria Between the Two Micro-Climatic Divergent Slopes at "Evolution Canyon" I, Israel.Front Genet. 2018 Nov 14;9:506. doi: 10.3389/fgene.2018.00506. eCollection 2018. Front Genet. 2018. PMID: 30487810 Free PMC article.
-
Genome-Wide Association Study of Wood Anatomical and Morphological Traits in Populus trichocarpa.Front Plant Sci. 2020 Sep 9;11:545748. doi: 10.3389/fpls.2020.545748. eCollection 2020. Front Plant Sci. 2020. PMID: 33013968 Free PMC article.
-
Identification of a novel abscisic acid-regulated farnesol dehydrogenase from Arabidopsis.Plant Physiol. 2010 Nov;154(3):1116-27. doi: 10.1104/pp.110.157784. Epub 2010 Aug 31. Plant Physiol. 2010. PMID: 20807998 Free PMC article.
References
-
- Abell BM, High S, Moloney MM. Membrane protein topology of oleosin is constrained by its long hydrophobic domain. J. Biol. Chem. 2002;277:8602–8610. - PubMed
-
- Baker A, Sparkes IA. Peroxisome protein import: some answers, more questions. Curr. Opin. Plant Biol. 2005;8:640–647. - PubMed
-
- Beigneux A, Withycombe SK, Digits JA, Tschantz WR, Weinbaum CA, Griffey SM, Bergo M, Casey PJ, Young SG. Prenylcysteine lyase deficiency in mice results in the accumulation of farnesylcysteine and geranylgeranylcysteine in brain and liver. J. Biol. Chem. 2002;277:38358–38363. - PubMed
-
- Bentinger M, Grunler J, Peterson E, Swiezewska E, Dallner G. Phosphorylation of farnesol in rat liver microsomes: properties of farnesol kinase and farnesyl phosphate kinase. Arch. Biochem. Biophys. 1998;353:191–198. - PubMed
-
- Bergo MO, Leung GK, Ambroziak P, Otto JC, Casey PJ, Gomes AQ, Seabra MC, Young SG. Isoprenylcysteine carboxyl methyltransferase deficiency in mice. J. Biol. Chem. 2001;276:5841–5845. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

