Chain and conformation stability of solid-state DNA: implications for room temperature storage
- PMID: 19969539
- PMCID: PMC2836546
- DOI: 10.1093/nar/gkp1060
Chain and conformation stability of solid-state DNA: implications for room temperature storage
Abstract
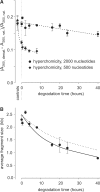
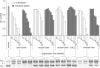
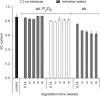
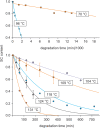
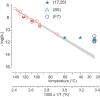
There is currently wide interest in room temperature storage of dehydrated DNA. However, there is insufficient knowledge about its chemical and structural stability. Here, we show that solid-state DNA degradation is greatly affected by atmospheric water and oxygen at room temperature. In these conditions DNA can even be lost by aggregation. These are major concerns since laboratory plastic ware is not airtight. Chain-breaking rates measured between 70 degrees C and 140 degrees C seemed to follow Arrhenius' law. Extrapolation to 25 degrees C gave a degradation rate of about 1-40 cuts/10(5) nucleotides/century. However, these figures are to be taken as very tentative since they depend on the validity of the extrapolation and the positive or negative effect of contaminants, buffers or additives. Regarding the secondary structure, denaturation experiments showed that DNA secondary structure could be preserved or fully restored upon rehydration, except possibly for small fragments. Indeed, below about 500 bp, DNA fragments underwent a very slow evolution (almost suppressed in the presence of trehalose) which could end in an irreversible denaturation. Thus, this work validates using room temperature for storage of DNA if completely protected from water and oxygen.
Figures

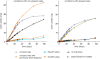






References
-
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. - PubMed
-
- Mitchell D, Willerslev E, Hansen A. Damage and repair of ancient DNA. Mutat. Res. 2005;571:265–276. - PubMed
-
- Pogocki D, Schoneich C. Chemical stability of nucleic acid-derived drugs. J. Pharm. Sci. 2000;89:443–456. - PubMed
-
- Pogozelski WK, Tullius TD. Oxidative strand scission of nucleic acids: routes initiated by hydrogen abstraction from the sugar moiety. Chem. Rev. 1998;98:1089–1108. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

