Selective inhibition of DNA replicase assembly by a non-natural nucleotide: exploiting the structural diversity of ATP-binding sites
- PMID: 19994907
- PMCID: PMC3169429
- DOI: 10.1021/cb900218c
Selective inhibition of DNA replicase assembly by a non-natural nucleotide: exploiting the structural diversity of ATP-binding sites
Abstract
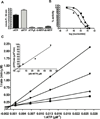
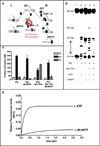
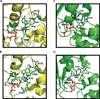
DNA synthesis is catalyzed by an ensemble of proteins designated the replicase. The efficient assembly of this multiprotein complex is essential for the continuity of DNA replication and is mediated by clamp-loading accessory proteins that use ATP binding and hydrolysis to coordinate these events. As a consequence, the ability to selectively inhibit the activity of these accessory proteins provides a rational approach to regulate DNA synthesis. Toward this goal, we tested the ability of several non-natural nucleotides to inhibit ATP-dependent enzymes associated with DNA replicase assembly. Kinetic and biophysical studies identified 5-nitro-indolyl-2'-deoxyribose-5'-triphosphate as a unique non-natural nucleotide capable of selectively inhibiting the bacteriophage T4 clamp loader versus the homologous enzyme from Escherichia coli. Modeling studies highlight the structural diversity between the ATP-binding site of each enzyme and provide a mechanism accounting for the differences in potencies for various substituted indolyl-2'-deoxyribose-5'-triphosphates. An in vivo assay measuring plaque formation demonstrates the efficacy and selectivity of 5-nitro-indolyl-2'-deoxyribose as a cytostatic agent against T4 bacteriophage while leaving viability of the E. coli host unaffected. This strategy provides a novel approach to develop agents that selectively inhibit ATP-dependent enzymes that are required for efficient DNA replication.
Figures

Similar articles
-
Dissection of the ATP-driven reaction cycle of the bacteriophage T4 DNA replication processivity clamp loading system.J Mol Biol. 2001 Jun 15;309(4):869-91. doi: 10.1006/jmbi.2001.4687. J Mol Biol. 2001. PMID: 11399065
-
An alternative clamp loading pathway via the T4 clamp loader gp44/62-DNA complex.Biochemistry. 2006 Jul 4;45(26):7976-89. doi: 10.1021/bi0601205. Biochemistry. 2006. PMID: 16800623
-
How a DNA polymerase clamp loader opens a sliding clamp.Science. 2011 Dec 23;334(6063):1675-80. doi: 10.1126/science.1211884. Science. 2011. PMID: 22194570 Free PMC article.
-
Opening of the clamp: an intimate view of an ATP-driven biological machine.Cell. 2001 Sep 21;106(6):655-60. doi: 10.1016/s0092-8674(01)00498-6. Cell. 2001. PMID: 11572772 Review.
-
Intricacies in ATP-dependent clamp loading: variations across replication systems.Structure. 2001 Nov;9(11):999-1004. doi: 10.1016/s0969-2126(01)00676-1. Structure. 2001. PMID: 11709164 Review.
Cited by
-
Evaluating the therapeutic potential of a non-natural nucleotide that inhibits human ribonucleotide reductase.Mol Cancer Ther. 2012 Oct;11(10):2077-86. doi: 10.1158/1535-7163.MCT-12-0199. Epub 2012 Aug 28. Mol Cancer Ther. 2012. PMID: 22933704 Free PMC article.
-
Architecture and conservation of the bacterial DNA replication machinery, an underexploited drug target.Curr Drug Targets. 2012 Mar;13(3):352-72. doi: 10.2174/138945012799424598. Curr Drug Targets. 2012. PMID: 22206257 Free PMC article. Review.
-
Synthetic nucleotides as probes of DNA polymerase specificity.J Nucleic Acids. 2012;2012:530963. doi: 10.1155/2012/530963. Epub 2012 Jun 7. J Nucleic Acids. 2012. PMID: 22720133 Free PMC article.
-
A novel non-natural nucleoside that influences P-glycoprotein activity and mediates drug resistance.Biochemistry. 2010 Mar 2;49(8):1640-8. doi: 10.1021/bi9020428. Biochemistry. 2010. PMID: 20104904 Free PMC article.
References
-
- Gerson SL. Clinical relevance of MGMT in the treatment of cancer. J Clin Oncol. 2002;20(9):2388–2399. - PubMed
-
- David Golan ATJ, Ehrin Armstrong, Joshua Galanter, April Armstrong, Ramy Arnaout, Harris Rose. Principles of Pharmacology: The Pathophysiologic Basis of Drug Therapy. Philidephia: Lippincott Williams & Wilkens; 2005.
-
- Laghi L, Bianchi P, Malesci A. Differences and evolution of the methods for the assessment of microsatellite instability. Oncogene. 2008;27(49):6313–6321. - PubMed
-
- Vigouroux C, Gharakhanian S, Salhi Y, Nguyen TH, Adda N, Rozenbaum W, Capeau J. Adverse metabolic disorders during highly active antiretroviral treatments (HAART) of HIV disease. Diabetes Metab. 1999;25(5):383–392. - PubMed
-
- Allan JM, Travis LB. Mechanisms of therapy-related carcinogenesis. Nat Rev Cancer. 2005;5(12):943–955. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Chemical Information
Research Materials

