The pitfalls of platform comparison: DNA copy number array technologies assessed
- PMID: 19995423
- PMCID: PMC2797821
- DOI: 10.1186/1471-2164-10-588
The pitfalls of platform comparison: DNA copy number array technologies assessed
Abstract
Background: The accurate and high resolution mapping of DNA copy number aberrations has become an important tool by which to gain insight into the mechanisms of tumourigenesis. There are various commercially available platforms for such studies, but there remains no general consensus as to the optimal platform. There have been several previous platform comparison studies, but they have either described older technologies, used less-complex samples, or have not addressed the issue of the inherent biases in such comparisons. Here we describe a systematic comparison of data from four leading microarray technologies (the Affymetrix Genome-wide SNP 5.0 array, Agilent High-Density CGH Human 244A array, Illumina HumanCNV370-Duo DNA Analysis BeadChip, and the Nimblegen 385 K oligonucleotide array). We compare samples derived from primary breast tumours and their corresponding matched normals, well-established cancer cell lines, and HapMap individuals. By careful consideration and avoidance of potential sources of bias, we aim to provide a fair assessment of platform performance.
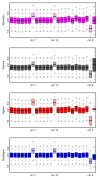
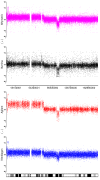
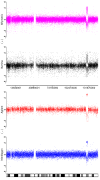
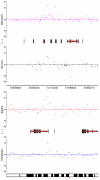
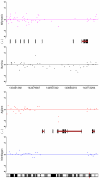
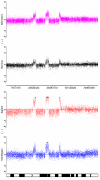
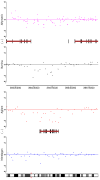
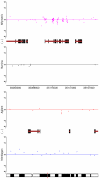
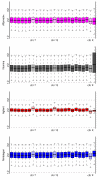
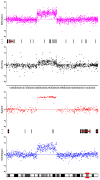
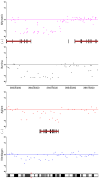
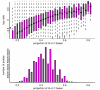
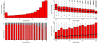
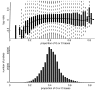
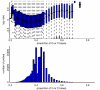
Results: By performing a theoretical assessment of the reproducibility, noise, and sensitivity of each platform, notable differences were revealed. Nimblegen exhibited between-replicate array variances an order of magnitude greater than the other three platforms, with Agilent slightly outperforming the others, and a comparison of self-self hybridizations revealed similar patterns. An assessment of the single probe power revealed that Agilent exhibits the highest sensitivity. Additionally, we performed an in-depth visual assessment of the ability of each platform to detect aberrations of varying sizes. As expected, all platforms were able to identify large aberrations in a robust manner. However, some focal amplifications and deletions were only detected in a subset of the platforms.
Conclusion: Although there are substantial differences in the design, density, and number of replicate probes, the comparison indicates a generally high level of concordance between platforms, despite differences in the reproducibility, noise, and sensitivity. In general, Agilent tended to be the best aCGH platform and Affymetrix, the superior SNP-CGH platform, but for specific decisions the results described herein provide a guide for platform selection and study design, and the dataset a resource for more tailored comparisons.
Figures


References
-
- Gaasenbeek M, Howarth K, Rowan AJ, Gorman PA, Jones A, Chaplin T, Liu Y, Bicknell D, Davison EJ, Fiegler H. Combined array-comparative genomic hybridization and single-nucleotide polymorphism-loss of heterozygosity analysis reveals complex changes and multiple forms of chromosomal instability in colorectal cancers. Cancer Res. 2006;66(7):3471–3479. doi: 10.1158/0008-5472.CAN-05-3285. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

