Cytosolic N-terminal arginine-based signals together with a luminal signal target a type II membrane protein to the plant ER
- PMID: 19995436
- PMCID: PMC2799409
- DOI: 10.1186/1471-2229-9-144
Cytosolic N-terminal arginine-based signals together with a luminal signal target a type II membrane protein to the plant ER
Abstract
Background: In eukaryotic cells, the membrane compartments that constitute the exocytic pathway are traversed by a constant flow of lipids and proteins. This is particularly true for the endoplasmic reticulum (ER), the main "gateway of the secretory pathway", where biosynthesis of sterols, lipids, membrane-bound and soluble proteins, and glycoproteins occurs. Maintenance of the resident proteins in this compartment implies they have to be distinguished from the secretory cargo. To this end, they must possess specific ER localization determinants to prevent their exit from the ER, and/or to interact with receptors responsible for their retrieval from the Golgi apparatus. Very few information is available about the signal(s) involved in the retention of membrane type II protein in the ER but it is generally accepted that sorting of ER type II cargo membrane proteins depends on motifs mainly located in their cytosolic tails.
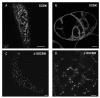
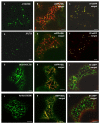
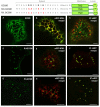
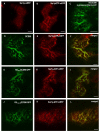
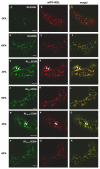
Results: Here, using Arabidopsis glucosidase I as a model, we have identified two types of signals sufficient for the location of a type II membrane protein in the ER. A first signal is located in the luminal domain, while a second signal corresponds to a short amino acid sequence located in the cytosolic tail of the membrane protein. The cytosolic tail contains at its N-terminal end four arginine residues constitutive of three di-arginine motifs (RR, RXR or RXXR) independently sufficient to confer ER localization. Interestingly, when only one di-arginine motif is present, fusion proteins are located both in the ER and in mobile punctate structures, distinct but close to Golgi bodies. Soluble and membrane ER protein markers are excluded from these punctate structures, which also do not colocalize with an ER-exit-site marker. It is hypothesized they correspond to sites involved in Golgi to ER retrotransport.
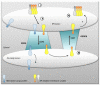
Conclusion: Altogether, these results clearly show that cytosolic and luminal signals responsible for ER retention could coexist in a same type II membrane protein. These data also suggest that both retrieval and retention mechanisms govern protein residency in the ER membrane. We hypothesized that mobile punctate structures not yet described at the ER/Golgi interface and tentatively named GERES, could be involved in retrieval mechanisms from the Golgi to the ER.
Figures




References
-
- Napier RM, Fowke LC, Hawes C, Lewis M, Pelham HR. Immunological evidence that plants use both HDEL and KDEL for targeting proteins to the endoplasmic reticulum. J Cell Sci. 1992;102:261–271. - PubMed
-
- Gomord V, Denmat LA, Fitchette-Lainé AC, Satiat-Jeunemaitre B, Hawes C, Faye L. The C-terminal HDEL sequence is sufficient for retention of secretory proteins in the endoplasmic reticulum (ER) but promotes vacuolar targeting of proteins that escape the ER. Plant J. 1997;11:313–325. doi: 10.1046/j.1365-313X.1997.11020313.x. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

