Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading
- PMID: 19996087
- PMCID: PMC2813476
- DOI: 10.1101/gr.097873.109
Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading
Abstract
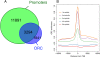
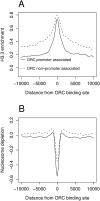
The origin recognition complex (ORC) is an essential DNA replication initiation factor conserved in all eukaryotes. In Saccharomyces cerevisiae, ORC binds to specific DNA elements; however, in higher eukaryotes, ORC exhibits little sequence specificity in vitro or in vivo. We investigated the genome-wide distribution of ORC in Drosophila and found that ORC localizes to specific chromosomal locations in the absence of any discernible simple motif. Although no clear sequence motif emerged, we were able to use machine learning approaches to accurately discriminate between ORC-associated sequences and ORC-free sequences based solely on primary sequence. The complex sequence features that define ORC binding sites are highly correlated with nucleosome positioning signals and likely represent a preferred nucleosomal landscape for ORC association. Open chromatin appears to be the underlying feature that is deterministic for ORC binding. ORC-associated sequences are enriched for the histone variant, H3.3, often at transcription start sites, and depleted for bulk nucleosomes. The density of ORC binding along the chromosome is reflected in the time at which a sequence replicates, with early replicating sequences having a high density of ORC binding. Finally, we found a high concordance between sites of ORC binding and cohesin loading, suggesting that, in addition to DNA replication, ORC may be required for the loading of cohesin on DNA in Drosophila.
Figures
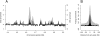
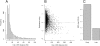
Similar articles
-
[Ability of Su(Hw) to create a platform for ORC binding does not depend on the type of surrounding chromatin].Tsitologiia. 2013;55(4):218-24. Tsitologiia. 2013. PMID: 23875451 Russian.
-
Forced binding of the origin of replication complex to chromosomal sites in Drosophila S2 cells creates an origin of replication.J Cell Sci. 2012 Feb 15;125(Pt 4):965-72. doi: 10.1242/jcs.094409. Epub 2012 Mar 15. J Cell Sci. 2012. PMID: 22421364
-
Insulator protein Su(Hw) recruits SAGA and Brahma complexes and constitutes part of Origin Recognition Complex-binding sites in the Drosophila genome.Nucleic Acids Res. 2013 Jun;41(11):5717-30. doi: 10.1093/nar/gkt297. Epub 2013 Apr 22. Nucleic Acids Res. 2013. PMID: 23609538 Free PMC article.
-
Changing the DNA landscape: putting a SPN on chromatin.Curr Top Microbiol Immunol. 2003;274:171-201. doi: 10.1007/978-3-642-55747-7_7. Curr Top Microbiol Immunol. 2003. PMID: 12596908 Review.
-
Multiple functions of the origin recognition complex.Int Rev Cytol. 2007;256:69-109. doi: 10.1016/S0074-7696(07)56003-1. Int Rev Cytol. 2007. PMID: 17241905 Review.
Cited by
-
Regulating DNA replication in plants.Cold Spring Harb Perspect Biol. 2012 Dec 1;4(12):a010140. doi: 10.1101/cshperspect.a010140. Cold Spring Harb Perspect Biol. 2012. PMID: 23209151 Free PMC article. Review.
-
Functional Coupling between DNA Replication and Sister Chromatid Cohesion Establishment.Int J Mol Sci. 2021 Mar 10;22(6):2810. doi: 10.3390/ijms22062810. Int J Mol Sci. 2021. PMID: 33802105 Free PMC article. Review.
-
Characterizing and controlling intrinsic biases of lambda exonuclease in nascent strand sequencing reveals phasing between nucleosomes and G-quadruplex motifs around a subset of human replication origins.Genome Res. 2015 May;25(5):725-35. doi: 10.1101/gr.183848.114. Epub 2015 Feb 18. Genome Res. 2015. PMID: 25695952 Free PMC article.
-
Genomic variation in natural populations of Drosophila melanogaster.Genetics. 2012 Oct;192(2):533-98. doi: 10.1534/genetics.112.142018. Epub 2012 Jun 5. Genetics. 2012. PMID: 22673804 Free PMC article.
-
Regulation of DNA replication by chromatin structures: accessibility and recruitment.Chromosoma. 2011 Feb;120(1):39-46. doi: 10.1007/s00412-010-0287-4. Epub 2010 Aug 3. Chromosoma. 2011. PMID: 20680317 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
