Regulation of gonadotropin-releasing hormone-1 gene transcription by members of the purine-rich element-binding protein family
- PMID: 19996387
- PMCID: PMC2838525
- DOI: 10.1152/ajpendo.00597.2009
Regulation of gonadotropin-releasing hormone-1 gene transcription by members of the purine-rich element-binding protein family
Abstract
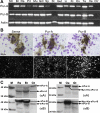
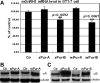
Gonadotropin-releasing hormone-1 (GnRH1) controls reproduction by stimulating the release of gonadotropins from the pituitary. To characterize regulatory factors governing GnRH1 gene expression, we employed biochemical and bioinformatics techniques to identify novel GnRH1 promoter-binding proteins from the brain of the cichlid fish, Astatotilapia burtoni (A. burtoni). Using an in vitro DNA-binding assay followed by mass spectrometric peptide mapping, we identified two members of the purine-rich element-binding (Pur) protein family, Puralpha and Purbeta, as candidates for GnRH1 promoter binding and regulation. We found that transcripts for both Puralpha and Purbeta colocalize in GnRH1-expressing neurons in the preoptic area of the hypothalamus in A. burtoni brain. Furthermore, we confirmed in vivo binding of endogenous Puralpha and Purbeta to the upstream region of the GnRH1 gene in A. burtoni brain and mouse neuronal GT1-7 cells. Consistent with the relative promoter occupancy exhibited by endogenous Pur proteins, overexpression of Purbeta, but not Puralpha, significantly downregulated GnRH1 mRNA levels in transiently transfected GT1-7 cells, suggesting that Purbeta acts as a repressor of GnRH1 gene transcription.
Figures


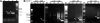

Similar articles
-
Heterogeneous nuclear ribonucleoprotein A/B and G inhibits the transcription of gonadotropin-releasing-hormone 1.Mol Cell Neurosci. 2008 Jan;37(1):69-84. doi: 10.1016/j.mcn.2007.08.015. Epub 2007 Aug 29. Mol Cell Neurosci. 2008. PMID: 17920292 Free PMC article.
-
Electrical synapses connect a network of gonadotropin releasing hormone neurons in a cichlid fish.Proc Natl Acad Sci U S A. 2015 Mar 24;112(12):3805-10. doi: 10.1073/pnas.1421851112. Epub 2015 Mar 9. Proc Natl Acad Sci U S A. 2015. PMID: 25775522 Free PMC article.
-
Luteinizing hormone beta promoter stimulation by adenylyl cyclase and cooperation with gonadotropin-releasing hormone 1 in transgenic mice and LBetaT2 Cells.Biol Reprod. 2007 Dec;77(6):1073-80. doi: 10.1095/biolreprod.107.064139. Epub 2007 Aug 15. Biol Reprod. 2007. PMID: 17699734
-
Mechanisms for pulsatile regulation of the gonadotropin subunit genes by GNRH1.Biol Reprod. 2006 Jun;74(6):993-8. doi: 10.1095/biolreprod.105.049049. Epub 2006 Feb 15. Biol Reprod. 2006. PMID: 16481592 Review.
-
Photoperiod-dependent regulation of gonadotropin-releasing hormone 1 messenger ribonucleic acid levels in the songbird brain.Gen Comp Endocrinol. 2013 Sep 1;190:81-7. doi: 10.1016/j.ygcen.2013.04.011. Epub 2013 May 7. Gen Comp Endocrinol. 2013. PMID: 23660447 Free PMC article. Review.
Cited by
-
Social opportunity rapidly regulates expression of CRF and CRF receptors in the brain during social ascent of a teleost fish, Astatotilapia burtoni.PLoS One. 2014 May 13;9(5):e96632. doi: 10.1371/journal.pone.0096632. eCollection 2014. PLoS One. 2014. PMID: 24824619 Free PMC article.
-
Bpur, the Lyme disease spirochete's PUR domain protein: identification as a transcriptional modulator and characterization of nucleic acid interactions.J Biol Chem. 2013 Sep 6;288(36):26220-26234. doi: 10.1074/jbc.M113.491357. Epub 2013 Jul 11. J Biol Chem. 2013. PMID: 23846702 Free PMC article.
-
Whole exome sequencing in family trios reveals de novo mutations in PURA as a cause of severe neurodevelopmental delay and learning disability.J Med Genet. 2014 Dec;51(12):806-13. doi: 10.1136/jmedgenet-2014-102798. Epub 2014 Oct 23. J Med Genet. 2014. PMID: 25342064 Free PMC article.
-
Gonadotropin-releasing hormone plasticity: a comparative perspective.Front Neuroendocrinol. 2012 Aug;33(3):287-300. doi: 10.1016/j.yfrne.2012.09.001. Epub 2012 Oct 3. Front Neuroendocrinol. 2012. PMID: 23041619 Free PMC article. Review.
References
-
- Belsham DD, Lovejoy DA. Gonadotropin-releasing hormone: gene evolution, expression, and regulation. Vitam Horm 71: 59–94, 2005 - PubMed
-
- Blake CA, Brown LM, Duncan MW, Hunsucker SW, Helmke SM. Estrogen regulation of the rat anterior pituitary gland proteome. Exp Biol Med (Maywood) 230: 800–807, 2005 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

