Genotyping of Trypanosoma cruzi: systematic selection of assays allowing rapid and accurate discrimination of all known lineages
- PMID: 19996435
- PMCID: PMC2825677
- DOI: 10.4269/ajtmh.2009.09-0305
Genotyping of Trypanosoma cruzi: systematic selection of assays allowing rapid and accurate discrimination of all known lineages
Abstract
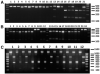
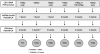
Trypanosoma cruzi, the agent of Chagas disease, can be subdivided into six discrete typing units (DTUs), TcI, TcIIa, TcIIb, TcIIc, TcIId or TcIIe, each having distinct epidemiologically important features. Dozens of genetic markers are available to determine the DTU to which a T. cruzi isolate belongs, but there is no consensus on which should be used. We selected five assays: three polymerase chain reaction (PCR)-restriction fragment length polymorphisms based on single nucleotide polymorphisms (SNPs) in the HSP60, Histone H1, and GPI loci, and PCR product size polymorphism of the LSU rDNA and mini-exon loci. Each assay was tested for its capacity to differentiate between DTUs using a panel of 48 genetically diverse T. cruzi clones. Some markers allowed unequivocal identification of individual DTUs, however, only by using a combination of multiple markers could all six DTUs be resolved. Based upon the results we recommend a triple-assay comprising the LSU rDNA, HSP60 and GPI markers for reliable, rapid, low-cost DTU assignment.
Figures

Similar articles
-
Design of a AFLP-PCR and PCR-RFLP test that identify the majority of discrete typing units of Trypanosoma cruzi.PLoS One. 2020 Aug 4;15(8):e0237180. doi: 10.1371/journal.pone.0237180. eCollection 2020. PLoS One. 2020. PMID: 32750094 Free PMC article.
-
Genotyping of Trypanosoma cruzi DTUs and Trypanosoma rangeli genetic groups in experimentally infected Rhodnius prolixus by PCR-RFLP.Acta Trop. 2016 Apr;156:115-21. doi: 10.1016/j.actatropica.2016.01.006. Epub 2016 Jan 11. Acta Trop. 2016. PMID: 26792202
-
Real-time PCR strategy for the identification of Trypanosoma cruzi discrete typing units directly in chronically infected human blood.Infect Genet Evol. 2017 Apr;49:300-308. doi: 10.1016/j.meegid.2017.02.006. Epub 2017 Feb 6. Infect Genet Evol. 2017. PMID: 28185987
-
Trypanosoma cruzi genetic diversity: Something new for something known about Chagas disease manifestations, serodiagnosis and drug sensitivity.Acta Trop. 2018 Aug;184:38-52. doi: 10.1016/j.actatropica.2017.09.017. Epub 2017 Sep 21. Acta Trop. 2018. PMID: 28941731 Review.
-
Trypanosoma cruzi I: Towards the need of genetic subdivision?, Part II.Acta Trop. 2018 Aug;184:53-58. doi: 10.1016/j.actatropica.2017.05.005. Epub 2017 May 8. Acta Trop. 2018. PMID: 28495405 Review.
Cited by
-
Wide reference databases for typing Trypanosoma cruzi based on amplicon sequencing of the minicircle hypervariable region.PLoS Negl Trop Dis. 2023 Nov 13;17(11):e0011764. doi: 10.1371/journal.pntd.0011764. eCollection 2023 Nov. PLoS Negl Trop Dis. 2023. PMID: 37956210 Free PMC article.
-
Between a bug and a hard place: Trypanosoma cruzi genetic diversity and the clinical outcomes of Chagas disease.Expert Rev Anti Infect Ther. 2015 Aug;13(8):995-1029. doi: 10.1586/14787210.2015.1056158. Expert Rev Anti Infect Ther. 2015. PMID: 26162928 Free PMC article. Review.
-
Multilocus sequence typing (MLST) for lineage assignment and high resolution diversity studies in Trypanosoma cruzi.PLoS Negl Trop Dis. 2011 Jun;5(6):e1049. doi: 10.1371/journal.pntd.0001049. Epub 2011 Jun 21. PLoS Negl Trop Dis. 2011. PMID: 21713026 Free PMC article.
-
Hosts and vectors of Trypanosoma cruzi discrete typing units in the Chagas disease endemic region of the Paraguayan Chaco.Parasitology. 2017 Jun;144(7):884-898. doi: 10.1017/S0031182016002663. Epub 2017 Feb 9. Parasitology. 2017. PMID: 28179034 Free PMC article.
-
New insights into Trypanosoma cruzi evolution, genotyping and molecular diagnostics from satellite DNA sequence analysis.PLoS Negl Trop Dis. 2017 Dec 18;11(12):e0006139. doi: 10.1371/journal.pntd.0006139. eCollection 2017 Dec. PLoS Negl Trop Dis. 2017. PMID: 29253860 Free PMC article.
References
-
- WHO . World Health Report. World Health Organisation; Geneva: 2002.
-
- Schofield CJ, Jannin J, Salvatella R. The future of Chagas disease control. Trends Parasitol. 2006;22:583–588. - PubMed
-
- Dias JC. Southern Cone Initiative for the elimination of domestic populations of Triatoma infestans and the interruption of transfusion Chagas disease: historical aspects, present situation, and perspectives. Mem Inst Oswaldo Cruz. 2007;102(Suppl 1):11–18. - PubMed
-
- Feliciangeli MD, Sanchez-Martin MJ, Suarez B, Marrero R, Torrellas A, Bravo A, Medina M, Martinez C, Hernandez M, Duque N, Toyo J, Rangel R. Risk factors for Trypanosoma cruzi human infection in Barinas State, Venezuela. Am J Trop Med Hyg. 2007;76:915–921. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

