Statistical tests of genetic association in the presence of gene-gene and gene-environment interactions
- PMID: 19996610
- PMCID: PMC2826874
- DOI: 10.1159/000264450
Statistical tests of genetic association in the presence of gene-gene and gene-environment interactions
Abstract
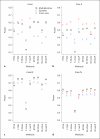
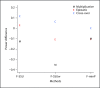
Background: While its importance is well recognized, it remains challenging to test genetic association in the presence of gene-gene (or gene-environment) interactions. A major technical difficulty lies in the fact that a general model of gene-gene interactions calls for the use of often a large number of parameters, leading to possibly reduced statistical power. An emerging theme of some recent work is to reduce the number of such parameters through dimension reduction. Wang et al. [2009] proposed such an approach based on the partial least squares (PLS) for dimension reduction. They compared their method with several others using simulated data, establishing that their PLS test performed best. Unfortunately, Wang et al. did not include in their evaluations several powerful tests just recently discovered for analyzing multiple SNPs in a candidate gene or region.
Methods: In this paper, we first extend these tests to the current context to detect gene-gene interactions in the presence of nuisance parameters, then compare these tests with the PLS test using the simulated data of Wang et al. [2009].
Results: It is confirmed that some other tests can be more powerful than the PLS test, though there is no uniform winner. Some interesting, albeit not new, observations are also made: some of the new tests are more robust to the large number of parameters in a model and may thus perform well; on the other hand, even for a purely epistatic genetic model, some of the tests applied to a logistic main-effects model without any interaction terms may be superior to that based on a full model that explicitly accounts for gene-gene interactions.
Conclusion: The proposed statistical tests are potentially useful in practice.
Copyright 2009 S. Karger AG, Basel.
Figures
Similar articles
-
Adaptive tests for detecting gene-gene and gene-environment interactions.Hum Hered. 2011;72(2):98-109. doi: 10.1159/000330632. Epub 2011 Sep 16. Hum Hered. 2011. PMID: 21934325 Free PMC article.
-
A partial least-square approach for modeling gene-gene and gene-environment interactions when multiple markers are genotyped.Genet Epidemiol. 2009 Jan;33(1):6-15. doi: 10.1002/gepi.20351. Genet Epidemiol. 2009. PMID: 18615621 Free PMC article.
-
A forest-based approach to identifying gene and gene gene interactions.Proc Natl Acad Sci U S A. 2007 Dec 4;104(49):19199-203. doi: 10.1073/pnas.0709868104. Epub 2007 Nov 28. Proc Natl Acad Sci U S A. 2007. PMID: 18048322 Free PMC article.
-
Analysis of Gene-Gene Interactions.Curr Protoc Hum Genet. 2017 Oct 18;95:1.14.1-1.14.10. doi: 10.1002/cphg.45. Curr Protoc Hum Genet. 2017. PMID: 29044470 Review.
-
[Posttraumatic stress disorder (PTSD) as a consequence of the interaction between an individual genetic susceptibility, a traumatogenic event and a social context].Encephale. 2012 Oct;38(5):373-80. doi: 10.1016/j.encep.2011.12.003. Epub 2012 Jan 24. Encephale. 2012. PMID: 23062450 Review. French.
Cited by
-
Detecting rare haplotype-environment interaction with logistic Bayesian LASSO.Genet Epidemiol. 2014 Jan;38(1):31-41. doi: 10.1002/gepi.21773. Epub 2013 Nov 23. Genet Epidemiol. 2014. PMID: 24272913 Free PMC article.
-
Adaptive tests for detecting gene-gene and gene-environment interactions.Hum Hered. 2011;72(2):98-109. doi: 10.1159/000330632. Epub 2011 Sep 16. Hum Hered. 2011. PMID: 21934325 Free PMC article.
-
Detecting a weak association by testing its multiple perturbations: a data mining approach.Sci Rep. 2014 May 28;4:5081. doi: 10.1038/srep05081. Sci Rep. 2014. PMID: 24866319 Free PMC article.
-
Powerful multi-marker association tests: unifying genomic distance-based regression and logistic regression.Genet Epidemiol. 2010 Nov;34(7):680-8. doi: 10.1002/gepi.20529. Genet Epidemiol. 2010. PMID: 20976795 Free PMC article.
-
Selecting Genetic Variants and Interactions Associated with Amyotrophic Lateral Sclerosis: A Group LASSO Approach.J Pers Med. 2022 Aug 19;12(8):1330. doi: 10.3390/jpm12081330. J Pers Med. 2022. PMID: 36013279 Free PMC article.
References
-
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. - PubMed
-
- Chatterjee N, Carroll RJ. Semiparametric maximum-likelihood estimation exploiting gene-environment independence in case-control studies. Biometrika. 2005;92:399–418.
-
- Clayton D, Chapman J, Cooper J. Use of unphased multilocus genotype data in indirect association studies. Genet Epidemiol. 2004;27:415–428. - PubMed
-
- Cox DR, Hinkley DV: Theoretical Statistics, Chapman and Hall, London.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources