APOBEC3G expression is dysregulated in primary HIV-1 infection and polymorphic variants influence CD4+ T-cell counts and plasma viral load
- PMID: 19996938
- PMCID: PMC3470914
- DOI: 10.1097/QAD.0b013e3283353bba
APOBEC3G expression is dysregulated in primary HIV-1 infection and polymorphic variants influence CD4+ T-cell counts and plasma viral load
Abstract
Objectives: In the absence of HIV-1 virion infectivity factor (Vif), cellular cytosine deaminases such as apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G (APOBEC3G) inhibit the virus by inducing hypermutations on viral DNA, among other mechanisms of action. We investigated the association of APOBEC3G mRNA levels and genetic variants on HIV-1 susceptibility, and early disease pathogenesis using viral load and CD4 T-cell counts as outcomes.
Methods: Study participants were 250 South African women at high risk for HIV-1 subtype C infection. We used real-time PCR to measure the expression of APOBEC3G in HIV-negative and HIV-positive primary infection samples. APOBEC3G variants were identified by DNA re-sequencing and TaqMan genotyping.
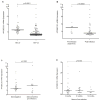
Results: We found no correlation between APOBEC3G expression levels and plasma viral loads (r = 0.053, P = 0.596) or CD4 T-cell counts (r = 0.030, P = 0.762) in 32 seroconverters. APOBEC3G expression levels were higher in HIV-negative individuals as compared with HIV-positive individuals (P < 0.0001), including matched pre and postinfection samples from the same individuals (n = 13, P < 0.0001). Twenty-four single nucleotide polymorphisms, including eight novel, were identified within APOBEC3G by re-sequencing and genotyping. The H186R mutation, a codon-changing variant in exon 4, and a 3' extragenic mutation (rs35228531) were associated with high viral loads (P = 0.0097 and P < 0.0001) and decreased CD4 T-cell levels (P = 0.0081 and P < 0.0001), respectively.
Conclusion: These data suggest that APOBEC3G transcription is rapidly downregulated upon HIV-1 infection. During primary infection, APOBEC3G expression levels in peripheral blood mononuclear cells do not correlate with viral loads or CD4 T-cell counts. Genetic variation of APOBEC3G may significantly affect early HIV-1 pathogenesis, although the mechanism remains unclear and warrants further investigation.
Figures

Similar articles
-
APOBEC3G mRNA expression in exposed seronegative and early stage HIV infected individuals decreases with removal of exposure and with disease progression.Retrovirology. 2009 Mar 2;6:23. doi: 10.1186/1742-4690-6-23. Retrovirology. 2009. PMID: 19254362 Free PMC article.
-
APOBEC3G/3A Expression in Human Immunodeficiency Virus Type 1-Infected Individuals Following Initiation of Antiretroviral Therapy Containing Cenicriviroc or Efavirenz.Front Immunol. 2018 Aug 8;9:1839. doi: 10.3389/fimmu.2018.01839. eCollection 2018. Front Immunol. 2018. PMID: 30135687 Free PMC article.
-
Possible footprints of APOBEC3F and/or other APOBEC3 deaminases, but not APOBEC3G, on HIV-1 from patients with acute/early and chronic infections.J Virol. 2014 Nov;88(21):12882-94. doi: 10.1128/JVI.01460-14. Epub 2014 Aug 27. J Virol. 2014. PMID: 25165112 Free PMC article.
-
[Advances in the study of molecular mechanism of APOBEC3G anti-HIV-1].Yao Xue Xue Bao. 2008 Jul;43(7):678-82. Yao Xue Xue Bao. 2008. PMID: 18819469 Review. Chinese.
-
Protecting APOBEC3G: a potential new target for HIV drug discovery.Curr Opin Investig Drugs. 2005 Feb;6(2):141-7. Curr Opin Investig Drugs. 2005. PMID: 15751736 Review.
Cited by
-
The KT Jeang retrovirology prize 2023: Thumbi Ndung'u.Retrovirology. 2023 Oct 18;20(1):17. doi: 10.1186/s12977-023-00632-9. Retrovirology. 2023. PMID: 37848918 Free PMC article. No abstract available.
-
HIV-1 competition experiments in humanized mice show that APOBEC3H imposes selective pressure and promotes virus adaptation.PLoS Pathog. 2017 May 5;13(5):e1006348. doi: 10.1371/journal.ppat.1006348. eCollection 2017 May. PLoS Pathog. 2017. PMID: 28475648 Free PMC article.
-
Natural Resistance to HIV Infection: Role of Immune Activation.Immun Inflamm Dis. 2025 Feb;13(2):e70138. doi: 10.1002/iid3.70138. Immun Inflamm Dis. 2025. PMID: 39998960 Free PMC article. Review.
-
The effect of HIV-1 Vif polymorphisms on A3G anti-viral activity in an in vivo mouse model.Retrovirology. 2016 Jun 30;13(1):45. doi: 10.1186/s12977-016-0280-y. Retrovirology. 2016. PMID: 27363431 Free PMC article.
-
Host Factors and HIV-1 Replication: Clinical Evidence and Potential Therapeutic Approaches.Front Immunol. 2013 Oct 24;4:343. doi: 10.3389/fimmu.2013.00343. Front Immunol. 2013. PMID: 24167505 Free PMC article. Review.
References
-
- Huthoff H, Malim MH. Cytidine deamination and resistance to retroviral infection: towards a structural understanding of the APOBEC proteins. Virology. 2005;334:147–153. - PubMed
-
- Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat Rev Immunol. 2004;4:868–877. - PubMed
-
- Bishop KN, Holmes RK, Sheehy AM, Davidson NO, Cho SJ, Malim MH. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr Biol. 2004;14:1392–1396. - PubMed
-
- Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. - PubMed
-
- Stephen JO, Nelson GW. Human genes that limit AIDS. Nature Genetics. 2004;36:565–574. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

