Origin and evolution of the adaptive immune system: genetic events and selective pressures
- PMID: 19997068
- PMCID: PMC3805090
- DOI: 10.1038/nrg2703
Origin and evolution of the adaptive immune system: genetic events and selective pressures
Abstract
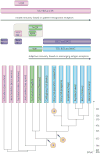
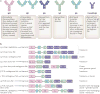
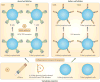
The adaptive immune system (AIS) in mammals, which is centred on lymphocytes bearing antigen receptors that are generated by somatic recombination, arose approximately 500 million years ago in jawed fish. This intricate defence system consists of many molecules, mechanisms and tissues that are not present in jawless vertebrates. Two macroevolutionary events are believed to have contributed to the genesis of the AIS: the emergence of the recombination-activating gene (RAG) transposon, and two rounds of whole-genome duplication. It has recently been discovered that a non-RAG-based AIS with similarities to the jawed vertebrate AIS - including two lymphoid cell lineages - arose in jawless fish by convergent evolution. We offer insights into the latest advances in this field and speculate on the selective pressures that led to the emergence and maintenance of the AIS.
Figures

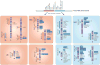
References
-
- Weigert MG, Cesari IM, Yonkovich SJ, Cohn M. Variability in the λ light chain sequences of mouse antibody. Nature. 1970;228:1045–1047. - PubMed
-
- Davis MM, Chien YH, Gascoigne NR, Hedrick SM. A murine T cell receptor gene complex: isolation, structure and rearrangement. Immunol Rev. 1984;81:235–258. - PubMed
-
- Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. - PubMed
-
- Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

