The origin recognition complex interacts with a subset of metabolic genes tightly linked to origins of replication
- PMID: 19997491
- PMCID: PMC2778871
- DOI: 10.1371/journal.pgen.1000755
The origin recognition complex interacts with a subset of metabolic genes tightly linked to origins of replication
Abstract
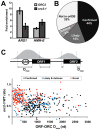
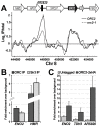
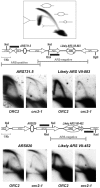
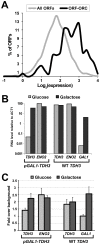
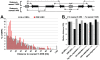
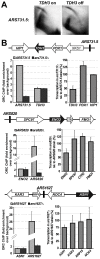
The origin recognition complex (ORC) marks chromosomal sites as replication origins and is essential for replication initiation. In yeast, ORC also binds to DNA elements called silencers, where its primary function is to recruit silent information regulator (SIR) proteins to establish transcriptional silencing. Indeed, silencers function poorly as chromosomal origins. Several genetic, molecular, and biochemical studies of HMR-E have led to a model proposing that when ORC becomes limiting in the cell (such as in the orc2-1 mutant) only sites that bind ORC tightly (such as HMR-E) remain fully occupied by ORC, while lower affinity sites, including many origins, lose ORC occupancy. Since HMR-E possessed a unique non-replication function, we reasoned that other tight sites might reveal novel functions for ORC on chromosomes. Therefore, we comprehensively determined ORC "affinity" genome-wide by performing an ORC ChIP-on-chip in ORC2 and orc2-1 strains. Here we describe a novel group of orc2-1-resistant ORC-interacting chromosomal sites (ORF-ORC sites) that did not function as replication origins or silencers. Instead, ORF-ORC sites were comprised of protein-coding regions of highly transcribed metabolic genes. In contrast to the ORC-silencer paradigm, transcriptional activation promoted ORC association with these genes. Remarkably, ORF-ORC genes were enriched in proximity to origins of replication and, in several instances, were transcriptionally regulated by these origins. Taken together, these results suggest a surprising connection among ORC, replication origins, and cellular metabolism.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
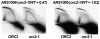
Similar articles
-
A role for a replicator dominance mechanism in silencing.EMBO J. 1999 Jul 1;18(13):3808-19. doi: 10.1093/emboj/18.13.3808. EMBO J. 1999. PMID: 10393196 Free PMC article.
-
The conserved bromo-adjacent homology domain of yeast Orc1 functions in the selection of DNA replication origins within chromatin.Genes Dev. 2010 Jul 1;24(13):1418-33. doi: 10.1101/gad.1906410. Genes Dev. 2010. PMID: 20595233 Free PMC article.
-
Functions of protosilencers in the formation and maintenance of heterochromatin in Saccharomyces cerevisiae.PLoS One. 2012;7(5):e37092. doi: 10.1371/journal.pone.0037092. Epub 2012 May 17. PLoS One. 2012. PMID: 22615905 Free PMC article.
-
Stepwise assembly of initiation complexes at budding yeast replication origins during the cell cycle.J Cell Sci Suppl. 1995;19:67-72. doi: 10.1242/jcs.1995.supplement_19.9. J Cell Sci Suppl. 1995. PMID: 8655649 Review.
-
A revisionist replicon model for higher eukaryotic genomes.J Cell Biochem. 2008 Oct 1;105(2):321-9. doi: 10.1002/jcb.21828. J Cell Biochem. 2008. PMID: 18680119 Free PMC article. Review.
Cited by
-
Design of a minimal silencer for the silent mating-type locus HML of Saccharomyces cerevisiae.Nucleic Acids Res. 2010 Dec;38(22):7991-8000. doi: 10.1093/nar/gkq689. Epub 2010 Aug 10. Nucleic Acids Res. 2010. PMID: 20699276 Free PMC article.
-
Mechanisms and regulation of DNA replication initiation in eukaryotes.Crit Rev Biochem Mol Biol. 2017 Apr;52(2):107-144. doi: 10.1080/10409238.2016.1274717. Epub 2017 Jan 17. Crit Rev Biochem Mol Biol. 2017. PMID: 28094588 Free PMC article. Review.
-
Mathematical modeling of genome replication.Phys Rev E Stat Nonlin Soft Matter Phys. 2012 Sep;86(3 Pt 1):031916. doi: 10.1103/PhysRevE.86.031916. Epub 2012 Sep 17. Phys Rev E Stat Nonlin Soft Matter Phys. 2012. PMID: 23030953 Free PMC article.
-
Mathematical modelling of whole chromosome replication.Nucleic Acids Res. 2010 Sep;38(17):5623-33. doi: 10.1093/nar/gkq343. Epub 2010 May 10. Nucleic Acids Res. 2010. PMID: 20457753 Free PMC article.
-
Origin replication complex binding, nucleosome depletion patterns, and a primary sequence motif can predict origins of replication in a genome with epigenetic centromeres.mBio. 2014 Sep 2;5(5):e01703-14. doi: 10.1128/mBio.01703-14. mBio. 2014. PMID: 25182328 Free PMC article.
References
-
- Bell SP. The origin recognition complex: from simple origins to complex functions. Genes Dev. 2002;16:659–72. - PubMed
-
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–74. - PubMed
-
- Jorgensen P, Tyers M. How cells coordinate growth and division. Curr Biol. 2004;14:R1014–27. - PubMed
-
- Mori S, Shirahige K. Perturbation of the activity of replication origin by meiosis-specific transcription. J Biol Chem. 2007;282:4447–52. - PubMed
-
- Friedman KL, Brewer BJ, Fangman WL. Replication profile of Saccharomyces cerevisiae chromosome VI. Genes Cells. 1997;2:667–78. - PubMed

