Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa
- PMID: 19997497
- PMCID: PMC2779102
- DOI: 10.1371/journal.pgen.1000753
Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa
Abstract
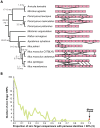
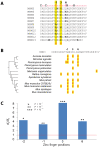
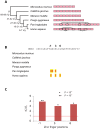
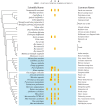
The onset of prezygotic and postzygotic barriers to gene flow between populations is a hallmark of speciation. One of the earliest postzygotic isolating barriers to arise between incipient species is the sterility of the heterogametic sex in interspecies' hybrids. Four genes that underlie hybrid sterility have been identified in animals: Odysseus, JYalpha, and Overdrive in Drosophila and Prdm9 (Meisetz) in mice. Mouse Prdm9 encodes a protein with a KRAB motif, a histone methyltransferase domain and several zinc fingers. The difference of a single zinc finger distinguishes Prdm9 alleles that cause hybrid sterility from those that do not. We find that concerted evolution and positive selection have rapidly altered the number and sequence of Prdm9 zinc fingers across 13 rodent genomes. The patterns of positive selection in Prdm9 zinc fingers imply that rapid evolution has acted on the interface between the Prdm9 protein and the DNA sequences to which it binds. Similar patterns are apparent for Prdm9 zinc fingers for diverse metazoans, including primates. Indeed, allelic variation at the DNA-binding positions of human PRDM9 zinc fingers show significant association with decreased risk of infertility. Prdm9 thus plays a role in determining male sterility both between species (mouse) and within species (human). The recurrent episodes of positive selection acting on Prdm9 suggest that the DNA sequences to which it binds must also be evolving rapidly. Our findings do not identify the nature of the underlying DNA sequences, but argue against the proposed role of Prdm9 as an essential transcription factor in mouse meiosis. We propose a hypothetical model in which incompatibilities between Prdm9-binding specificity and satellite DNAs provide the molecular basis for Prdm9-mediated hybrid sterility. We suggest that Prdm9 should be investigated as a candidate gene in other instances of hybrid sterility in metazoans.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Muller HJ. Isolating mechanisms, evolution, and temperature. Biol Symp. 1942;6:71–125.
-
- Coyne J, Orr HA. Speciation: Sinauer Associates, Sunderland MA 2004
-
- Orr HA, Turelli M. The evolution of postzygotic isolation: accumulating Dobzhansky-Muller incompatibilities. Evolution. 2001;55:1085–1094. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

