Investigation of the genes involved in antigenic switching at the vlsE locus in Borrelia burgdorferi: an essential role for the RuvAB branch migrase
- PMID: 19997508
- PMCID: PMC2779866
- DOI: 10.1371/journal.ppat.1000680
Investigation of the genes involved in antigenic switching at the vlsE locus in Borrelia burgdorferi: an essential role for the RuvAB branch migrase
Abstract
Persistent infection by pathogenic organisms requires effective strategies for the defense of these organisms against the host immune response. A common strategy employed by many pathogens to escape immune recognition and clearance is to continually vary surface epitopes through recombinational shuffling of genetic information. Borrelia burgdorferi, a causative agent of Lyme borreliosis, encodes a surface-bound lipoprotein, VlsE. This protein is encoded by the vlsE locus carried at the right end of the linear plasmid lp28-1. Adjacent to the expression locus are 15 silent cassettes carrying information that is moved into the vlsE locus through segmental gene conversion events. The protein players and molecular mechanism of recombinational switching at vlsE have not been characterized. In this study, we analyzed the effect of the independent disruption of 17 genes that encode factors involved in DNA recombination, repair or replication on recombinational switching at the vlsE locus during murine infection. In Neisseria gonorrhoeae, 10 such genes have been implicated in recombinational switching at the pilE locus. Eight of these genes, including recA, are either absent from B. burgdorferi, or do not show an obvious requirement for switching at vlsE. The only genes that are required in both organisms are ruvA and ruvB, which encode subunits of a Holliday junction branch migrase. Disruption of these genes results in a dramatic decrease in vlsE recombination with a phenotype similar to that observed for lp28-1 or vls-minus spirochetes: productive infection at week 1 with clearance by day 21. In SCID mice, the persistence defect observed with ruvA and ruvB mutants was fully rescued as previously observed for vlsE-deficient B. burgdorferi. We report the requirement of the RuvAB branch migrase in recombinational switching at vlsE, the first essential factor to be identified in this process. These findings are supported by the independent work of Lin et al. in the accompanying article, who also found a requirement for the RuvAB branch migrase. Our results also indicate that the mechanism of switching at vlsE in B. burgdorferi is distinct from switching at pilE in N. gonorrhoeae, which is the only other organism analyzed genetically in detail. Finally, our findings suggest a unique mechanism for switching at vlsE and a role for currently unidentified B. burgdorferi proteins in this process.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
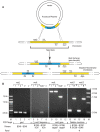

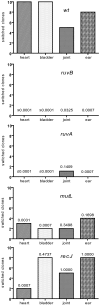
Similar articles
-
vls Antigenic Variation Systems of Lyme Disease Borrelia: Eluding Host Immunity through both Random, Segmental Gene Conversion and Framework Heterogeneity.Microbiol Spectr. 2014 Dec;2(6):10.1128/microbiolspec.MDNA3-0038-2014. doi: 10.1128/microbiolspec.MDNA3-0038-2014. Microbiol Spectr. 2014. PMID: 26104445 Free PMC article. Review.
-
Central role of the Holliday junction helicase RuvAB in vlsE recombination and infectivity of Borrelia burgdorferi.PLoS Pathog. 2009 Dec;5(12):e1000679. doi: 10.1371/journal.ppat.1000679. Epub 2009 Dec 4. PLoS Pathog. 2009. PMID: 19997622 Free PMC article.
-
Antigenic variation in the Lyme spirochete: detailed functional assessment of recombinational switching at vlsE in the JD1 strain of Borrelia burgdorferi.Mol Microbiol. 2019 Mar;111(3):750-763. doi: 10.1111/mmi.14189. Epub 2019 Jan 31. Mol Microbiol. 2019. PMID: 30580501
-
Detailed analysis of sequence changes occurring during vlsE antigenic variation in the mouse model of Borrelia burgdorferi infection.PLoS Pathog. 2009 Feb;5(2):e1000293. doi: 10.1371/journal.ppat.1000293. Epub 2009 Feb 13. PLoS Pathog. 2009. PMID: 19214205 Free PMC article.
-
Changing of the guard: How the Lyme disease spirochete subverts the host immune response.J Biol Chem. 2020 Jan 10;295(2):301-313. doi: 10.1074/jbc.REV119.008583. Epub 2019 Nov 21. J Biol Chem. 2020. PMID: 31753921 Free PMC article. Review.
Cited by
-
YebC regulates variable surface antigen VlsE expression and is required for host immune evasion in Borrelia burgdorferi.PLoS Pathog. 2020 Oct 13;16(10):e1008953. doi: 10.1371/journal.ppat.1008953. eCollection 2020 Oct. PLoS Pathog. 2020. PMID: 33048986 Free PMC article.
-
Mini-review: Strategies for Variation and Evolution of Bacterial Antigens.Comput Struct Biotechnol J. 2015 Jul 26;13:407-16. doi: 10.1016/j.csbj.2015.07.002. eCollection 2015. Comput Struct Biotechnol J. 2015. PMID: 26288700 Free PMC article. Review.
-
Genetic Manipulation of Borrelia Spp.Curr Top Microbiol Immunol. 2018;415:113-140. doi: 10.1007/82_2017_51. Curr Top Microbiol Immunol. 2018. PMID: 28918538 Free PMC article. Review.
-
Lyme Disease Pathogenesis.Curr Issues Mol Biol. 2021;42:473-518. doi: 10.21775/cimb.042.473. Epub 2020 Dec 23. Curr Issues Mol Biol. 2021. PMID: 33353871 Free PMC article. Review.
-
vls Antigenic Variation Systems of Lyme Disease Borrelia: Eluding Host Immunity through both Random, Segmental Gene Conversion and Framework Heterogeneity.Microbiol Spectr. 2014 Dec;2(6):10.1128/microbiolspec.MDNA3-0038-2014. doi: 10.1128/microbiolspec.MDNA3-0038-2014. Microbiol Spectr. 2014. PMID: 26104445 Free PMC article. Review.
References
-
- Dzikowski R, Deitsch K. Antigenic variation by protozoan parasites: insights from Babesia bovis. Mol Microbiol. 2006;59:364–366. - PubMed
-
- Dzikowski R, Templeton TJ, Deitsch K. Variant antigen gene expression in malaria. Cell Microbiol. 2006;8:1371–1381. - PubMed
-
- Taylor JE, Rudenko G. Switching trypanosome coats: what's in the wardrobe? Trends Genet. 2006;22:614–620. - PubMed
-
- Brayton KA, Palmer GH, Lundgren A, Yi J, Barbet AF. Antigenic variation of Anaplasma marginale msp2 occurs by combinatorial gene conversion. Mol Microbiol. 2002;43:1151–1159. - PubMed
-
- Palmer GH, Brayton KA. Gene conversion is a convergent strategy for pathogen antigenic variation. Trends Parasitol. 2007;23:408–413. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

