Multi-organ expression profiling uncovers a gene module in coronary artery disease involving transendothelial migration of leukocytes and LIM domain binding 2: the Stockholm Atherosclerosis Gene Expression (STAGE) study
- PMID: 19997623
- PMCID: PMC2780352
- DOI: 10.1371/journal.pgen.1000754
Multi-organ expression profiling uncovers a gene module in coronary artery disease involving transendothelial migration of leukocytes and LIM domain binding 2: the Stockholm Atherosclerosis Gene Expression (STAGE) study
Abstract
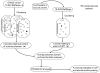
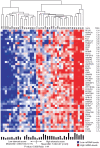
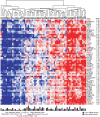
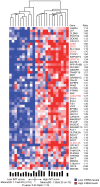
Environmental exposures filtered through the genetic make-up of each individual alter the transcriptional repertoire in organs central to metabolic homeostasis, thereby affecting arterial lipid accumulation, inflammation, and the development of coronary artery disease (CAD). The primary aim of the Stockholm Atherosclerosis Gene Expression (STAGE) study was to determine whether there are functionally associated genes (rather than individual genes) important for CAD development. To this end, two-way clustering was used on 278 transcriptional profiles of liver, skeletal muscle, and visceral fat (n = 66/tissue) and atherosclerotic and unaffected arterial wall (n = 40/tissue) isolated from CAD patients during coronary artery bypass surgery. The first step, across all mRNA signals (n = 15,042/12,621 RefSeqs/genes) in each tissue, resulted in a total of 60 tissue clusters (n = 3958 genes). In the second step (performed within tissue clusters), one atherosclerotic lesion (n = 49/48) and one visceral fat (n = 59) cluster segregated the patients into two groups that differed in the extent of coronary stenosis (P = 0.008 and P = 0.00015). The associations of these clusters with coronary atherosclerosis were validated by analyzing carotid atherosclerosis expression profiles. Remarkably, in one cluster (n = 55/54) relating to carotid stenosis (P = 0.04), 27 genes in the two clusters relating to coronary stenosis were confirmed (n = 16/17, P<10(-27 and-30)). Genes in the transendothelial migration of leukocytes (TEML) pathway were overrepresented in all three clusters, referred to as the atherosclerosis module (A-module). In a second validation step, using three independent cohorts, the A-module was found to be genetically enriched with CAD risk by 1.8-fold (P<0.004). The transcription co-factor LIM domain binding 2 (LDB2) was identified as a potential high-hierarchy regulator of the A-module, a notion supported by subnetwork analysis, by cellular and lesion expression of LDB2, and by the expression of 13 TEML genes in Ldb2-deficient arterial wall. Thus, the A-module appears to be important for atherosclerosis development and, together with LDB2, merits further attention in CAD research.
Conflict of interest statement
Clinical Gene Networks AB with Johan Björkegren and Jesper Tegnér as major shareholders has filed a PCT application for a screening method using genes in the atherosclerosis module including LDB2 (PCT/SE2007/00864).
Figures

References
-
- Ginsburg GS, Donahue MP, Newby LK. Prospects for personalized cardiovascular medicine: the impact of genomics. J Am Coll Cardiol. 2005;46:1615–1627. - PubMed
-
- Schadt EE, Sachs A, Friend S. Embracing complexity, inching closer to reality. Sci STKE. 2005;2005:pe40. - PubMed
-
- Tegner J, Skogsberg J, Bjorkegren J. Thematic review series: systems biology approaches to metabolic and cardiovascular disorders. Multi-organ whole-genome measurements and reverse engineering to uncover gene networks underlying complex traits. J Lipid Res. 2007;48:267–277. - PubMed
-
- Blatt M, Wiseman S, Domany E. Superparamagnetic clustering of data. Phys Rev Lett. 1996;76:3251–3254. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

