NMR analysis of the dynamic exchange of the NS2B cofactor between open and closed conformations of the West Nile virus NS2B-NS3 protease
- PMID: 19997625
- PMCID: PMC2780355
- DOI: 10.1371/journal.pntd.0000561
NMR analysis of the dynamic exchange of the NS2B cofactor between open and closed conformations of the West Nile virus NS2B-NS3 protease
Abstract
Background: The two-component NS2B-NS3 proteases of West Nile and dengue viruses are essential for viral replication and established targets for drug development. In all crystal structures of the proteases to date, the NS2B cofactor is located far from the substrate binding site (open conformation) in the absence of inhibitor and lining the substrate binding site (closed conformation) in the presence of an inhibitor.
Methods: In this work, nuclear magnetic resonance (NMR) spectroscopy of isotope and spin-labeled samples of the West Nile virus protease was used to investigate the occurrence of equilibria between open and closed conformations in solution.
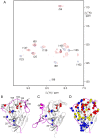
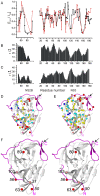
Findings: In solution, the closed form of the West Nile virus protease is the predominant conformation irrespective of the presence or absence of inhibitors. Nonetheless, dissociation of the C-terminal part of the NS2B cofactor from the NS3 protease (open conformation) occurs in both the presence and the absence of inhibitors. Low-molecular-weight inhibitors can shift the conformational exchange equilibria so that over 90% of the West Nile virus protease molecules assume the closed conformation. The West Nile virus protease differs from the dengue virus protease, where the open conformation is the predominant form in the absence of inhibitors.
Conclusion: Partial dissociation of NS2B from NS3 has implications for the way in which the NS3 protease can be positioned with respect to the host cell membrane when NS2B is membrane associated via N- and C-terminal segments present in the polyprotein. In the case of the West Nile virus protease, discovery of low-molecular-weight inhibitors that act by breaking the association of the NS2B cofactor with the NS3 protease is impeded by the natural affinity of the cofactor to the NS3 protease. The same strategy can be more successful in the case of the dengue virus NS2B-NS3 protease.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Binding of low molecular weight inhibitors promotes large conformational changes in the dengue virus NS2B-NS3 protease: fold analysis by pseudocontact shifts.J Am Chem Soc. 2011 Nov 30;133(47):19205-15. doi: 10.1021/ja208435s. Epub 2011 Nov 9. J Am Chem Soc. 2011. PMID: 22007671
-
Profiling of flaviviral NS2B-NS3 protease specificity provides a structural basis for the development of selective chemical tools that differentiate Dengue from Zika and West Nile viruses.Antiviral Res. 2020 Mar;175:104731. doi: 10.1016/j.antiviral.2020.104731. Epub 2020 Jan 31. Antiviral Res. 2020. PMID: 32014497
-
Homology modeling and molecular dynamics study of West Nile virus NS3 protease: a molecular basis for the catalytic activity increased by the NS2B cofactor.Proteins. 2006 Nov 15;65(3):692-701. doi: 10.1002/prot.21129. Proteins. 2006. PMID: 16972281
-
West Nile Virus NS2B/NS3 protease as an antiviral target.Curr Med Chem. 2008;15(27):2771-84. doi: 10.2174/092986708786242804. Curr Med Chem. 2008. PMID: 18991636 Review.
-
The Structure of the Zika Virus Protease, NS2B/NS3pro.Adv Exp Med Biol. 2018;1062:131-145. doi: 10.1007/978-981-10-8727-1_10. Adv Exp Med Biol. 2018. PMID: 29845530 Review.
Cited by
-
Proteases from dengue, West Nile and Zika viruses as drug targets.Biophys Rev. 2019 Apr;11(2):157-165. doi: 10.1007/s12551-019-00508-3. Epub 2019 Feb 26. Biophys Rev. 2019. PMID: 30806881 Free PMC article. Review.
-
A conformational switch high-throughput screening assay and allosteric inhibition of the flavivirus NS2B-NS3 protease.PLoS Pathog. 2017 May 25;13(5):e1006411. doi: 10.1371/journal.ppat.1006411. eCollection 2017 May. PLoS Pathog. 2017. PMID: 28542603 Free PMC article.
-
Potential Dual Role of West Nile Virus NS2B in Orchestrating NS3 Enzymatic Activity in Viral Replication.Viruses. 2021 Jan 31;13(2):216. doi: 10.3390/v13020216. Viruses. 2021. PMID: 33572517 Free PMC article.
-
Identifying Allosteric Small-Molecule Binding Sites of Inactive NS2B-NS3 Proteases of Pathogenic Flaviviridae.Viruses. 2024 Dec 24;17(1):6. doi: 10.3390/v17010006. Viruses. 2024. PMID: 39861795 Free PMC article.
-
Dengue virus protease activity modulated by dynamics of protease cofactor.Biophys J. 2021 Jun 15;120(12):2444-2453. doi: 10.1016/j.bpj.2021.04.015. Epub 2021 Apr 22. Biophys J. 2021. PMID: 33894215 Free PMC article.
References
-
- Lanciotti RS, Roehrig JT, Deubel V, Smith J, Parker M, et al. Origin of the West Nile virus responsible for a outbreak of encephalitis in the Northeastern United States. Science. 1999;286:2333–2337. - PubMed
-
- Chappell KJ, Stoermer MJ, Fairlie DP, Young PR. Mutagenesis of the West Nile virus NS2B cofactor domain reveals two regions essential for protease activity. J Gen Virol. 2008;89:1010–1014. - PubMed
-
- Lescar J, Luo D, Xu T, Sampath A, Lim SP, et al. Towards the design of antiviral inhibitors against flaviviruses: the case for the multifunctional NS3 protein from Dengue virus as a target. Antiviral Res. 2008;80:94–101. - PubMed
-
- Stoermer MJ, Chappell KJ, Liebscher S, Jensen CM, Gan CH, et al. Potent cationic inhibitors of West Nile virus NS2B/NS3 protease with serum stability, cell permeability and antiviral activity. J Med Chem. 2008;51:5714–5721. - PubMed
-
- Chappell KJ, Stoermer MJ, Fairlie DP, Young PR. Generation and characterization of proteolytically active and highly stable truncated and full-length recombinant West Nile virus NS3. Prot Expr Purif. 2007;53:87–96. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

