Expression of genes encoding multi-transmembrane proteins in specific primate taste cell populations
- PMID: 19997627
- PMCID: PMC2780358
- DOI: 10.1371/journal.pone.0007682
Expression of genes encoding multi-transmembrane proteins in specific primate taste cell populations
Abstract
Background: Using fungiform (FG) and circumvallate (CV) taste buds isolated by laser capture microdissection and analyzed using gene arrays, we previously constructed a comprehensive database of gene expression in primates, which revealed over 2,300 taste bud-associated genes. Bioinformatics analyses identified hundreds of genes predicted to encode multi-transmembrane domain proteins with no previous association with taste function. A first step in elucidating the roles these gene products play in gustation is to identify the specific taste cell types in which they are expressed.
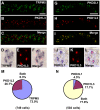
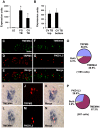
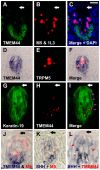
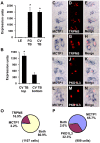
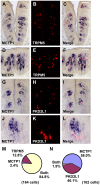
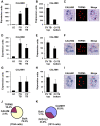
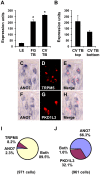
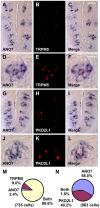
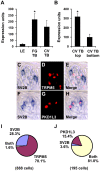
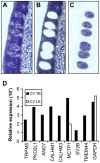
Methodology/principal findings: Using double label in situ hybridization analyses, we identified seven new genes expressed in specific taste cell types, including sweet, bitter, and umami cells (TRPM5-positive), sour cells (PKD2L1-positive), as well as other taste cell populations. Transmembrane protein 44 (TMEM44), a protein with seven predicted transmembrane domains with no homology to GPCRs, is expressed in a TRPM5-negative and PKD2L1-negative population that is enriched in the bottom portion of taste buds and may represent developmentally immature taste cells. Calcium homeostasis modulator 1 (CALHM1), a component of a novel calcium channel, along with family members CALHM2 and CALHM3; multiple C2 domains; transmembrane 1 (MCTP1), a calcium-binding transmembrane protein; and anoctamin 7 (ANO7), a member of the recently identified calcium-gated chloride channel family, are all expressed in TRPM5 cells. These proteins may modulate and effect calcium signalling stemming from sweet, bitter, and umami receptor activation. Synaptic vesicle glycoprotein 2B (SV2B), a regulator of synaptic vesicle exocytosis, is expressed in PKD2L1 cells, suggesting that this taste cell population transmits tastant information to gustatory afferent nerve fibers via exocytic neurotransmitter release.
Conclusions/significance: Identification of genes encoding multi-transmembrane domain proteins expressed in primate taste buds provides new insights into the processes of taste cell development, signal transduction, and information coding. Discrete taste cell populations exhibit highly specific gene expression patterns, supporting a model whereby each mature taste receptor cell is responsible for sensing, transmitting, and coding a specific taste quality.
Conflict of interest statement
Figures
Similar articles
-
How do taste cells lacking synapses mediate neurotransmission? CALHM1, a voltage-gated ATP channel.Bioessays. 2013 Dec;35(12):1111-8. doi: 10.1002/bies.201300077. Epub 2013 Sep 17. Bioessays. 2013. PMID: 24105910 Free PMC article. Review.
-
Voltage-gated sodium channels in taste bud cells.BMC Neurosci. 2009 Mar 12;10:20. doi: 10.1186/1471-2202-10-20. BMC Neurosci. 2009. PMID: 19284629 Free PMC article.
-
Transient receptor potential family members PKD1L3 and PKD2L1 form a candidate sour taste receptor.Proc Natl Acad Sci U S A. 2006 Aug 15;103(33):12569-74. doi: 10.1073/pnas.0602702103. Epub 2006 Aug 4. Proc Natl Acad Sci U S A. 2006. PMID: 16891422 Free PMC article.
-
CALHM1 ion channel mediates purinergic neurotransmission of sweet, bitter and umami tastes.Nature. 2013 Mar 14;495(7440):223-6. doi: 10.1038/nature11906. Epub 2013 Mar 6. Nature. 2013. PMID: 23467090 Free PMC article.
-
Taste receptor signalling - from tongues to lungs.Acta Physiol (Oxf). 2012 Feb;204(2):158-68. doi: 10.1111/j.1748-1716.2011.02308.x. Epub 2011 May 7. Acta Physiol (Oxf). 2012. PMID: 21481196 Free PMC article. Review.
Cited by
-
ERK1/2 activation in human taste bud cells regulates fatty acid signaling and gustatory perception of fat in mice and humans.FASEB J. 2016 Oct;30(10):3489-3500. doi: 10.1096/fj.201600422R. Epub 2016 Jun 29. FASEB J. 2016. PMID: 27358389 Free PMC article.
-
Reduced expression of NGEP is associated with high-grade prostate cancers: a tissue microarray analysis.Cancer Immunol Immunother. 2013 Oct;62(10):1609-18. doi: 10.1007/s00262-013-1463-1. Epub 2013 Aug 17. Cancer Immunol Immunother. 2013. PMID: 23955683 Free PMC article.
-
Biology, function and structure of the calcium homeostasis modulator family.J Physiol. 2025 Aug;603(15):4179-4199. doi: 10.1113/JP285197. Epub 2024 Oct 29. J Physiol. 2025. PMID: 39470434 Review.
-
How do taste cells lacking synapses mediate neurotransmission? CALHM1, a voltage-gated ATP channel.Bioessays. 2013 Dec;35(12):1111-8. doi: 10.1002/bies.201300077. Epub 2013 Sep 17. Bioessays. 2013. PMID: 24105910 Free PMC article. Review.
-
On the molecular nature of large-pore channels.J Mol Biol. 2021 Aug 20;433(17):166994. doi: 10.1016/j.jmb.2021.166994. Epub 2021 Apr 16. J Mol Biol. 2021. PMID: 33865869 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

