Quantitative Correlation between the protein primary sequences and secondary structures in spider dragline silks
- PMID: 20000730
- PMCID: PMC2805410
- DOI: 10.1021/bm9010672
Quantitative Correlation between the protein primary sequences and secondary structures in spider dragline silks
Abstract
Synthetic spider silk holds great potential for use in various applications spanning medical uses to ultra lightweight armor; however, producing synthetic fibers with mechanical properties comparable to natural spider silk has eluded the scientific community. Natural dragline spider silks are commonly made from proteins that contain highly repetitive amino acid motifs, adopting an array of secondary structures. Before further advances can be made in the production of synthetic fibers based on spider silk proteins, it is imperative to know the percentage of each amino acid in the protein that forms a specific secondary structure. Linking these percentages to the primary amino acid sequence of the protein will establish a structural foundation for synthetic silk. In this study, nuclear magnetic resonance (NMR) techniques are used to quantify the percentage of Ala, Gly, and Ser that form both beta-sheet and helical secondary structures. The fraction of these three amino acids and their secondary structure are quantitatively correlated to the primary amino acid sequence for the proteins that comprise major and minor ampullate silk from the Nephila clavipes spider providing a blueprint for synthetic spider silks.
Figures
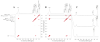
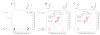
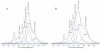



References
-
- Shear WA, Palmer JM, Coddington JA, Bonamo PM. Science. 1989;246:479–481. - PubMed
-
- Lewis R. Chem. Rev. 2006;106:3762–3774. - PubMed
-
- Vollrath F, Porter D. Soft Matter. 2006;2:377–385. - PubMed
-
- Gosline J, DeMont E, Denny M. Endeavour. 1986;10:37–43.
-
- Swanson BO, Blackledge TA, Beltran J, Hayashi CY. Appl. Phys. A. 2006;82:213–218.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

