'Unknown' proteins and 'orphan' enzymes: the missing half of the engineering parts list--and how to find it
- PMID: 20001958
- PMCID: PMC3022307
- DOI: 10.1042/BJ20091328
'Unknown' proteins and 'orphan' enzymes: the missing half of the engineering parts list--and how to find it
Abstract
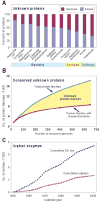
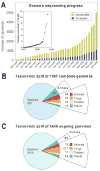
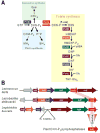
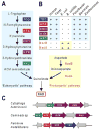
Like other forms of engineering, metabolic engineering requires knowledge of the components (the 'parts list') of the target system. Lack of such knowledge impairs both rational engineering design and diagnosis of the reasons for failures; it also poses problems for the related field of metabolic reconstruction, which uses a cell's parts list to recreate its metabolic activities in silico. Despite spectacular progress in genome sequencing, the parts lists for most organisms that we seek to manipulate remain highly incomplete, due to the dual problem of 'unknown' proteins and 'orphan' enzymes. The former are all the proteins deduced from genome sequence that have no known function, and the latter are all the enzymes described in the literature (and often catalogued in the EC database) for which no corresponding gene has been reported. Unknown proteins constitute up to about half of the proteins in prokaryotic genomes, and much more than this in higher plants and animals. Orphan enzymes make up more than a third of the EC database. Attacking the 'missing parts list' problem is accordingly one of the great challenges for post-genomic biology, and a tremendous opportunity to discover new facets of life's machinery. Success will require a co-ordinated community-wide attack, sustained over years. In this attack, comparative genomics is probably the single most effective strategy, for it can reliably predict functions for unknown proteins and genes for orphan enzymes. Furthermore, it is cost-efficient and increasingly straightforward to deploy owing to a proliferation of databases and associated tools.
Figures


Similar articles
-
Profiling the orphan enzymes.Biol Direct. 2014 Jun 6;9:10. doi: 10.1186/1745-6150-9-10. Biol Direct. 2014. PMID: 24906382 Free PMC article. Review.
-
An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review.
-
The CanOE strategy: integrating genomic and metabolic contexts across multiple prokaryote genomes to find candidate genes for orphan enzymes.PLoS Comput Biol. 2012 May;8(5):e1002540. doi: 10.1371/journal.pcbi.1002540. Epub 2012 May 31. PLoS Comput Biol. 2012. PMID: 22693442 Free PMC article.
-
Finding sequences for over 270 orphan enzymes.PLoS One. 2014 May 14;9(5):e97250. doi: 10.1371/journal.pone.0097250. eCollection 2014. PLoS One. 2014. PMID: 24826896 Free PMC article.
-
A survey of orphan enzyme activities.BMC Bioinformatics. 2007 Jul 10;8:244. doi: 10.1186/1471-2105-8-244. BMC Bioinformatics. 2007. PMID: 17623104 Free PMC article.
Cited by
-
Linking genome-scale metabolic modeling and genome annotation.Methods Mol Biol. 2013;985:61-83. doi: 10.1007/978-1-62703-299-5_4. Methods Mol Biol. 2013. PMID: 23417799 Free PMC article.
-
Identification of canine platelet proteins separated by differential detergent fractionation for nonelectrophoretic proteomics analyzed by Gene Ontology and pathways analysis.Vet Med (Auckl). 2014 Jun 21;5:1-9. doi: 10.2147/VMRR.S47127. eCollection 2014. Vet Med (Auckl). 2014. PMID: 32670841 Free PMC article.
-
Profiling the orphan enzymes.Biol Direct. 2014 Jun 6;9:10. doi: 10.1186/1745-6150-9-10. Biol Direct. 2014. PMID: 24906382 Free PMC article. Review.
-
Detecting anomalous proteins using deep representations.NAR Genom Bioinform. 2024 Feb 27;6(1):lqae021. doi: 10.1093/nargab/lqae021. eCollection 2024 Mar. NAR Genom Bioinform. 2024. PMID: 38486884 Free PMC article.
-
Functional Diversity of Haloacid Dehalogenase Superfamily Phosphatases from Saccharomyces cerevisiae: BIOCHEMICAL, STRUCTURAL, AND EVOLUTIONARY INSIGHTS.J Biol Chem. 2015 Jul 24;290(30):18678-98. doi: 10.1074/jbc.M115.657916. Epub 2015 Jun 12. J Biol Chem. 2015. PMID: 26071590 Free PMC article.
References
-
- Stephanopoulos GN, Aristidou AA, Nielsen J. Metabolic Engineering: Principles and Methodologies. Academic Press; San Diego: 1998.
-
- Hanson AD, Shanks JV. Plant metabolic engineering: entering the S curve. Metab Eng. 2002;4:1–2.
-
- Capell T, Christou P. Progress in plant metabolic engineering. Curr Opin Biotechnol. 2004;15:148–154. - PubMed
-
- Wu S, Chappell J. Metabolic engineering of natural products in plants; tools of the trade and challenges for the future. Curr Opin Biotechnol. 2008;19:145–152. - PubMed
-
- Kunze R, Frommer WB, Flügge UI. Metabolic engineering of plants: the role of membrane transport. Metab Eng. 2002;4:57–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

