Polyphosphate binds with high affinity to exosite II of thrombin
- PMID: 20002544
- PMCID: PMC2856763
- DOI: 10.1111/j.1538-7836.2009.03723.x
Polyphosphate binds with high affinity to exosite II of thrombin
Abstract
Background: Polyphosphate (a linear polymer of inorganic phosphate) is secreted from platelet dense granules, and we recently showed that it accelerates factor V activation by thrombin.
Objective: To examine the interaction of polyphosphate with thrombin.
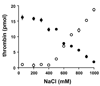
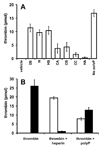
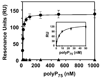
Methods and results: Thrombin, but not prothrombin, altered the electrophoretic migration of polyphosphate in gel mobility assays. Thrombin binding to polyphosphate was influenced by ionic strength, and was evident even in plasma. Two positively charged exosites on thrombin mediate its interactions with other proteins and accessory molecules: exosite I (mainly with thrombin substrates), and exosite II (mainly with certain anionic polymers). Free thrombin, thrombin in complex with hirudin's C-terminal dodecapeptide and gamma-thrombin all bound polyphosphate similarly, excluding exosite I involvement. Mutations within exosite II, but not within exosite I, the Na(+)-binding site or hydrophobic pocket, weakened thrombin binding to polyphosphate as revealed by NaCl dependence. Surface plasmon resonance demonstrated tight interaction of polyphosphate with thrombin (K(d) approximately 5 nm) but reduced interaction with a thrombin exosite II mutant. Certain glycosaminoglycans, including heparin, only partially competed with polyphosphate for binding to thrombin, and polyphosphate did not reduce heparin-catalyzed inactivation of thrombin by antithrombin.
Conclusion: Polyphosphate interacts with thrombin's exosite II at a site that partially overlaps with, but is not identical to, the heparin-binding site. Polyphosphate interactions with thrombin may be physiologically relevant, as the polyphosphate concentrations achievable following platelet activation are far above the approximately 5 nM K(d) for the polyphosphate-thrombin interaction.
Conflict of interest statement
N.J. Mutch and J.H. Morrissey are coinventors on pending patent applications covering some of the technologies discussed in this article.
Figures





Similar articles
-
Bothrojaracin: a potent two-site-directed thrombin inhibitor.Biochemistry. 1996 Jul 16;35(28):9083-9. doi: 10.1021/bi960043l. Biochemistry. 1996. PMID: 8703912
-
Interaction of thrombin with antithrombin, heparin cofactor II, and protein C inhibitor.J Protein Chem. 1993 Dec;12(6):677-88. doi: 10.1007/BF01024926. J Protein Chem. 1993. PMID: 8136018
-
Autoantibodies to thrombin directed against both of its cryptic exosites.Br J Haematol. 2006 Feb;132(4):487-93. doi: 10.1111/j.1365-2141.2005.05894.x. Br J Haematol. 2006. PMID: 16412021
-
Strategies for inhibiting the effects of thrombin.Blood Coagul Fibrinolysis. 1994 Jan;5 Suppl 1:S47-58; discussion S59-64. doi: 10.1097/00001721-199401000-00007. Blood Coagul Fibrinolysis. 1994. PMID: 8186356 Review.
-
A player of many parts: the spotlight falls on thrombin's structure.Thromb Res. 1993 Jan 1;69(1):1-58. doi: 10.1016/0049-3848(93)90002-6. Thromb Res. 1993. PMID: 8465268 Review.
Cited by
-
Positively-charged semi-tunnel is a structural and surface characteristic of polyphosphate-binding proteins: an in-silico study.PLoS One. 2015 Apr 16;10(4):e0123713. doi: 10.1371/journal.pone.0123713. eCollection 2015. PLoS One. 2015. PMID: 25879219 Free PMC article.
-
Thrombin-induced platelet activation via PAR4: pivotal role for exosite II.Thromb Haemost. 2014 Sep 2;112(3):558-65. doi: 10.1160/TH13-12-1013. Epub 2014 Jul 3. Thromb Haemost. 2014. PMID: 24990072 Free PMC article.
-
Polyphosphate Activates von Willebrand Factor Interaction with Glycoprotein Ib in the Absence of Factor VIII In Vitro.Int J Mol Sci. 2022 Nov 15;23(22):14118. doi: 10.3390/ijms232214118. Int J Mol Sci. 2022. PMID: 36430595 Free PMC article.
-
Polyphosphate multi-tasks.J Thromb Haemost. 2012 Nov;10(11):2313-4. doi: 10.1111/jth.12001. J Thromb Haemost. 2012. PMID: 23006797 Free PMC article. No abstract available.
-
A site on factor XII required for productive interactions with polyphosphate.J Thromb Haemost. 2023 Jun;21(6):1567-1579. doi: 10.1016/j.jtha.2023.02.014. Epub 2023 Mar 1. J Thromb Haemost. 2023. PMID: 36863563 Free PMC article.
References
-
- Naski MC, Shafer JA. Alpha-thrombin-catalyzed hydrolysis of fibrin I. Alternative binding modes and the accessibility of the active site in fibrin I-bound alpha-thrombin. J Biol Chem. 1990;265:1401–1407. - PubMed
-
- Tsiang M, Lentz SR, Dittman WA, Wen D, Scarpati EM, Sadler JE. Equilibrium binding of thrombin to recombinant human thrombomodulin: effect of hirudin, fibrinogen, factor Va, and peptide analogues. Biochemistry. 1990;29:10602–10612. - PubMed
-
- Rydel TJ, Tulinsky A, Bode W, Huber R. Refined structure of the hirudin-thrombin complex. J Mol Biol. 1991;221:583–601. - PubMed
-
- De Cristofaro R, De Candia E, Landolfi R, Rutella S, Hall SW. Structural and functional mapping of the thrombin domain involved in the binding to the platelet glycoprotein Ib. Biochemistry. 2001;40:13268–13273. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

