The hydrogenosomes of Psalteriomonas lanterna
- PMID: 20003182
- PMCID: PMC2796672
- DOI: 10.1186/1471-2148-9-287
The hydrogenosomes of Psalteriomonas lanterna
Abstract
Background: Hydrogenosomes are organelles that produce molecular hydrogen and ATP. The broad phylogenetic distribution of their hosts suggests that the hydrogenosomes of these organisms evolved several times independently from the mitochondria of aerobic progenitors. Morphology and 18S rRNA phylogeny suggest that the microaerophilic amoeboflagellate Psalteriomonas lanterna, which possesses hydrogenosomes and elusive "modified mitochondria", belongs to the Heterolobosea, a taxon that consists predominantly of aerobic, mitochondriate organisms. This taxon is rather unrelated to taxa with hitherto studied hydrogenosomes.
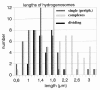
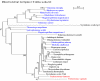
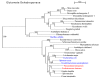
Results: Electron microscopy of P. lanterna flagellates reveals a large globule in the centre of the cell that is build up from stacks of some 20 individual hydrogenosomes. The individual hydrogenosomes are surrounded by a double membrane that encloses a homogeneous, dark staining matrix lacking cristae. The "modified mitochondria" are found in the cytoplasm of the cell and are surrounded by 1-2 cisterns of rough endoplasmatic reticulum, just as the mitochondria of certain related aerobic Heterolobosea. The ultrastructure of the "modified mitochondria" and hydrogenosomes is very similar, and they have the same size distribution as the hydrogenosomes that form the central stack.The phylogenetic analysis of selected EST sequences (Hsp60, Propionyl-CoA carboxylase) supports the phylogenetic position of P. lanterna close to aerobic Heterolobosea (Naegleria gruberi). Moreover, this analysis also confirms the identity of several mitochondrial or hydrogenosomal key-genes encoding proteins such as a Hsp60, a pyruvate:ferredoxin oxidoreductase, a putative ADP/ATP carrier, a mitochondrial complex I subunit (51 KDa), and a [FeFe] hydrogenase.
Conclusion: Comparison of the ultrastructure of the "modified mitochondria" and hydrogenosomes strongly suggests that both organelles are just two morphs of the same organelle. The EST studies suggest that the hydrogenosomes of P. lanterna are physiologically similar to the hydrogenosomes of Trichomonas vaginalis and Trimastix pyriformis. Phylogenetic analysis of the ESTs confirms the relationship of P. lanterna with its aerobic relative, the heterolobosean amoeboflagellate Naegleria gruberi, corroborating the evolution of hydrogenosomes from a common, mitochondriate ancestor.
Figures









Similar articles
-
Phylogenetic position of Psalteriomonas lanterna deduced from the SSU rDNA sequence.J Eukaryot Microbiol. 1997 Sep-Oct;44(5):467-70. doi: 10.1111/j.1550-7408.1997.tb05725.x. J Eukaryot Microbiol. 1997. PMID: 9304815
-
Genetic evidence for a mitochondriate ancestry in the 'amitochondriate' flagellate Trimastix pyriformis.PLoS One. 2008 Jan 2;3(1):e1383. doi: 10.1371/journal.pone.0001383. PLoS One. 2008. PMID: 18167542 Free PMC article.
-
Molecular cloning of hydrogenosomal ferredoxin cDNA from the anaerobic amoeboflagellate Psalteriomonas lanterna.Biochim Biophys Acta. 1994 Jan 4;1183(3):544-6. doi: 10.1016/0005-2728(94)90082-5. Biochim Biophys Acta. 1994. PMID: 8286402
-
Hydrogenosomes under microscopy.Tissue Cell. 2009 Jun;41(3):151-68. doi: 10.1016/j.tice.2009.01.001. Epub 2009 Mar 17. Tissue Cell. 2009. PMID: 19297000 Review.
-
Biogenesis of the hydrogenosome in the anaerobic protist Trichomonas vaginalis.J Parasitol. 1993 Oct;79(5):664-70. J Parasitol. 1993. PMID: 8410536 Review.
Cited by
-
Biochemistry and evolution of anaerobic energy metabolism in eukaryotes.Microbiol Mol Biol Rev. 2012 Jun;76(2):444-95. doi: 10.1128/MMBR.05024-11. Microbiol Mol Biol Rev. 2012. PMID: 22688819 Free PMC article. Review.
-
Energy metabolism in anaerobic eukaryotes and Earth's late oxygenation.Free Radic Biol Med. 2019 Aug 20;140:279-294. doi: 10.1016/j.freeradbiomed.2019.03.030. Epub 2019 Mar 29. Free Radic Biol Med. 2019. PMID: 30935869 Free PMC article. Review.
-
Adenine nucleotide transporters in organelles: novel genes and functions.Cell Mol Life Sci. 2011 Apr;68(7):1183-206. doi: 10.1007/s00018-010-0612-3. Epub 2011 Jan 5. Cell Mol Life Sci. 2011. PMID: 21207102 Free PMC article. Review.
-
The organellar genome and metabolic potential of the hydrogen-producing mitochondrion of Nyctotherus ovalis.Mol Biol Evol. 2011 Aug;28(8):2379-91. doi: 10.1093/molbev/msr059. Epub 2011 Mar 4. Mol Biol Evol. 2011. PMID: 21378103 Free PMC article.
-
The Naegleria genome: a free-living microbial eukaryote lends unique insights into core eukaryotic cell biology.Res Microbiol. 2011 Jul-Aug;162(6):607-18. doi: 10.1016/j.resmic.2011.03.003. Epub 2011 Mar 21. Res Microbiol. 2011. PMID: 21392573 Free PMC article. Review.
References
-
- Keithly J. In: Hydrogenosomes and mitosomes: mitochondria of anaerobic eukaryotes. Tachezy J, editor. Vol. 9. Berlin Heidelberg: Springer-Verlag; 2008. The mitochondrion related organelle of Cryptosporidium parvum; pp. 231–253. full_text.
-
- Muller M. The hydrogenosome. J Gen Microbiol. 1993;139(12):2879–2889. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

