Gene doctoring: a method for recombineering in laboratory and pathogenic Escherichia coli strains
- PMID: 20003185
- PMCID: PMC2796669
- DOI: 10.1186/1471-2180-9-252
Gene doctoring: a method for recombineering in laboratory and pathogenic Escherichia coli strains
Abstract
Background: Homologous recombination mediated by the lambda-Red genes is a common method for making chromosomal modifications in Escherichia coli. Several protocols have been developed that differ in the mechanisms by which DNA, carrying regions homologous to the chromosome, are delivered into the cell. A common technique is to electroporate linear DNA fragments into cells. Alternatively, DNA fragments are generated in vivo by digestion of a donor plasmid with a nuclease that does not cleave the host genome. In both cases the lambda-Red gene products recombine homologous regions carried on the linear DNA fragments with the chromosome. We have successfully used both techniques to generate chromosomal mutations in E. coli K-12 strains. However, we have had limited success with these lambda-Red based recombination techniques in pathogenic E. coli strains, which has led us to develop an enhanced protocol for recombineering in such strains.
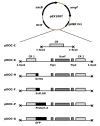
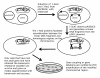
Results: Our goal was to develop a high-throughput recombineering system, primarily for the coupling of genes to epitope tags, which could also be used for deletion of genes in both pathogenic and K-12 E. coli strains. To that end we have designed a series of donor plasmids for use with the lambda-Red recombination system, which when cleaved in vivo by the I-SceI meganuclease generate a discrete linear DNA fragment, allowing for C-terminal tagging of chromosomal genes with a 6xHis, 3xFLAG, 4xProteinA or GFP tag or for the deletion of chromosomal regions. We have enhanced existing protocols and technologies by inclusion of a cassette conferring kanamycin resistance and, crucially, by including the sacB gene on the donor plasmid, so that all but true recombinants are counter-selected on kanamycin and sucrose containing media, thus eliminating the need for extensive screening. This method has the added advantage of limiting the exposure of cells to the potential damaging effects of the lambda-Red system, which can lead to unwanted secondary alterations to the chromosome.
Conclusion: We have developed a counter-selective recombineering technique for epitope tagging or for deleting genes in E. coli. We have demonstrated the versatility of the technique by modifying the chromosome of the enterohaemorrhagic O157:H7 (EHEC), uropathogenic CFT073 (UPEC), enteroaggregative O42 (EAEC) and enterotoxigenic H10407 (ETEC) E. coli strains as well as in K-12 laboratory strains.
Figures


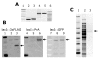
Similar articles
-
Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli.BMC Mol Biol. 2003 Dec 13;4:11. doi: 10.1186/1471-2199-4-11. BMC Mol Biol. 2003. PMID: 14672541 Free PMC article.
-
Recombineering: Genetic Engineering in Escherichia coli Using Homologous Recombination.Curr Protoc. 2023 Feb;3(2):e656. doi: 10.1002/cpz1.656. Curr Protoc. 2023. PMID: 36779782 Free PMC article.
-
Recombineering: genetic engineering in bacteria using homologous recombination.Curr Protoc Mol Biol. 2014 Apr 14;106:1.16.1-1.16.39. doi: 10.1002/0471142727.mb0116s106. Curr Protoc Mol Biol. 2014. PMID: 24733238 Review.
-
Golden Gate-Assisted Gene Doctoring for Streamlined and Efficient Recombineering in Bacteria.Methods Mol Biol. 2025;2850:345-363. doi: 10.1007/978-1-0716-4220-7_19. Methods Mol Biol. 2025. PMID: 39363081
-
λ Recombination and Recombineering.EcoSal Plus. 2016 May;7(1):10.1128/ecosalplus.ESP-0011-2015. doi: 10.1128/ecosalplus.ESP-0011-2015. EcoSal Plus. 2016. PMID: 27223821 Free PMC article. Review.
Cited by
-
A critical role for the cccA gene product, cytochrome c2, in diverting electrons from aerobic respiration to denitrification in Neisseria gonorrhoeae.J Bacteriol. 2013 Jun;195(11):2518-29. doi: 10.1128/JB.02300-12. Epub 2013 Mar 29. J Bacteriol. 2013. PMID: 23543713 Free PMC article.
-
All Three Endogenous Quinone Species of Escherichia coli Are Involved in Controlling the Activity of the Aerobic/Anaerobic Response Regulator ArcA.Front Microbiol. 2016 Sep 7;7:1339. doi: 10.3389/fmicb.2016.01339. eCollection 2016. Front Microbiol. 2016. PMID: 27656164 Free PMC article.
-
RNA polymerase supply and flux through the lac operon in Escherichia coli.Philos Trans R Soc Lond B Biol Sci. 2016 Nov 5;371(1707):20160080. doi: 10.1098/rstb.2016.0080. Philos Trans R Soc Lond B Biol Sci. 2016. PMID: 27672157 Free PMC article.
-
Detoxifying Escherichia coli for endotoxin-free production of recombinant proteins.Microb Cell Fact. 2015 Apr 16;14:57. doi: 10.1186/s12934-015-0241-5. Microb Cell Fact. 2015. PMID: 25890161 Free PMC article.
-
Isolation of generalized transducing bacteriophages for uropathogenic strains of Escherichia coli.Appl Environ Microbiol. 2011 Sep;77(18):6630-5. doi: 10.1128/AEM.05307-11. Epub 2011 Jul 22. Appl Environ Microbiol. 2011. PMID: 21784916 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

