The mitochondrial genomes of sponges provide evidence for multiple invasions by Repetitive Hairpin-forming Elements (RHE)
- PMID: 20003196
- PMCID: PMC2800124
- DOI: 10.1186/1471-2164-10-591
The mitochondrial genomes of sponges provide evidence for multiple invasions by Repetitive Hairpin-forming Elements (RHE)
Abstract
Background: The mitochondrial (mt) genomes of sponges possess a variety of features, which appear to be intermediate between those of Eumetazoa and non-metazoan opisthokonts. Among these features is the presence of long intergenic regions, which are common in other eukaryotes, but generally absent in Eumetazoa. Here we analyse poriferan mitochondrial intergenic regions, paying particular attention to repetitive sequences within them. In this context we introduce the mitochondrial genome of Ircinia strobilina (Lamarck, 1816; Demospongiae: Dictyoceratida) and compare it with mtDNA of other sponges.
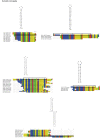
Results: Mt genomes of dictyoceratid sponges are identical in gene order and content but display major differences in size and organization of intergenic regions. An even higher degree of diversity in the structure of intergenic regions was found among different orders of demosponges. One interesting observation made from such comparisons was of what appears to be recurrent invasions of sponge mitochondrial genomes by repetitive hairpin-forming elements, which cause large genome size differences even among closely related taxa. These repetitive hairpin-forming elements are structurally and compositionally divergent and display a scattered distribution throughout various groups of demosponges.
Conclusion: Large intergenic regions of poriferan mt genomes are targets for insertions of repetitive hairpin- forming elements, similar to the ones found in non-metazoan opisthokonts. Such elements were likely present in some lineages early in animal mitochondrial genome evolution but were subsequently lost during the reduction of intergenic regions, which occurred in the Eumetazoa lineage after the split of Porifera. Porifera acquired their elements in several independent events. Patterns of their intra-genomic dispersal can be seen in the mt genome of Vaceletia sp.
Figures


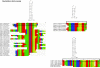

Similar articles
-
Seventeen new complete mtDNA sequences reveal extensive mitochondrial genome evolution within the Demospongiae.PLoS One. 2008 Jul 16;3(7):e2723. doi: 10.1371/journal.pone.0002723. PLoS One. 2008. PMID: 18628961 Free PMC article.
-
The complete mitochondrial genome of Ircinia sp. (Dictyoceratida: Irciniidae).Mitochondrial DNA. 2015 Apr;26(2):282-3. doi: 10.3109/19401736.2013.825776. Epub 2013 Sep 16. Mitochondrial DNA. 2015. PMID: 24041447
-
Mitochondrial genome of the homoscleromorph Oscarella carmela (Porifera, Demospongiae) reveals unexpected complexity in the common ancestor of sponges and other animals.Mol Biol Evol. 2007 Feb;24(2):363-73. doi: 10.1093/molbev/msl167. Epub 2006 Nov 7. Mol Biol Evol. 2007. PMID: 17090697
-
Deep phylogeny and evolution of sponges (phylum Porifera).Adv Mar Biol. 2012;61:1-78. doi: 10.1016/B978-0-12-387787-1.00007-6. Adv Mar Biol. 2012. PMID: 22560777 Review.
-
Comparative analyses of developmental transcription factor repertoires in sponges reveal unexpected complexity of the earliest animals.Mar Genomics. 2015 Dec;24 Pt 2:121-9. doi: 10.1016/j.margen.2015.07.008. Epub 2015 Aug 5. Mar Genomics. 2015. PMID: 26253310 Review.
Cited by
-
Cytonuclear Interactions in the Evolution of Animal Mitochondrial tRNA Metabolism.Genome Biol Evol. 2015 Jun 27;7(8):2089-101. doi: 10.1093/gbe/evv124. Genome Biol Evol. 2015. PMID: 26116918 Free PMC article.
-
Mitochondrial Genome Evolution of Placozoans: Gene Rearrangements and Repeat Expansions.Genome Biol Evol. 2021 Jan 7;13(1):evaa213. doi: 10.1093/gbe/evaa213. Genome Biol Evol. 2021. PMID: 33031489 Free PMC article.
-
Divergence times in demosponges (Porifera): first insights from new mitogenomes and the inclusion of fossils in a birth-death clock model.BMC Evol Biol. 2018 Jul 18;18(1):114. doi: 10.1186/s12862-018-1230-1. BMC Evol Biol. 2018. PMID: 30021516 Free PMC article.
-
The Skeleton Forming Proteome of an Early Branching Metazoan: A Molecular Survey of the Biomineralization Components Employed by the Coralline Sponge Vaceletia Sp.PLoS One. 2015 Nov 4;10(11):e0140100. doi: 10.1371/journal.pone.0140100. eCollection 2015. PLoS One. 2015. PMID: 26536128 Free PMC article.
-
Cnidarian phylogenetic relationships as revealed by mitogenomics.BMC Evol Biol. 2013 Jan 9;13:5. doi: 10.1186/1471-2148-13-5. BMC Evol Biol. 2013. PMID: 23302374 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

