Towards a comprehensive barcode library for arctic life - Ephemeroptera, Plecoptera, and Trichoptera of Churchill, Manitoba, Canada
- PMID: 20003245
- PMCID: PMC2800108
- DOI: 10.1186/1742-9994-6-30
Towards a comprehensive barcode library for arctic life - Ephemeroptera, Plecoptera, and Trichoptera of Churchill, Manitoba, Canada
Abstract
Background: This study reports progress in assembling a DNA barcode reference library for Ephemeroptera, Plecoptera, and Trichoptera ("EPTs") from a Canadian subarctic site, which is the focus of a comprehensive biodiversity inventory using DNA barcoding. These three groups of aquatic insects exhibit a moderate level of species diversity, making them ideal for testing the feasibility of DNA barcoding for routine biotic surveys. We explore the correlation between the morphological species delineations, DNA barcode-based haplotype clusters delimited by a sequence threshold (2%), and a threshold-free approach to biodiversity quantification--phylogenetic diversity.
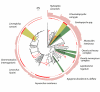
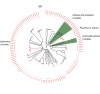
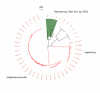
Results: A DNA barcode reference library is built for 112 EPT species for the focal region, consisting of 2277 COI sequences. Close correspondence was found between EPT morphospecies and haplotype clusters as designated using a standard threshold value. Similarly, the shapes of taxon accumulation curves based upon haplotype clusters were very similar to those generated using phylogenetic diversity accumulation curves, but were much more computationally efficient.
Conclusion: The results of this study will facilitate other lines of research on northern EPTs and also bode well for rapidly conducting initial biodiversity assessments in unknown EPT faunas.
Figures


References
-
- Wilson EO. The Diversity of Life. Cambridge: Harvard University Press; 1992.
-
- Ødegaard F. How many species of arthropods? Erwin's estimate revised. Biological Journal of the Linnean Society. 2000;71:583–597. doi: 10.1111/j.1095-8312.2000.tb01279.x. - DOI
-
- Gaston KJ, Blackburn TM. Pattern and processes in macroecology. Oxford: Blackwell Scientific; 2000.
LinkOut - more resources
Full Text Sources
Other Literature Sources

