CENPA overexpression promotes genome instability in pRb-depleted human cells
- PMID: 20003272
- PMCID: PMC2797498
- DOI: 10.1186/1476-4598-8-119
CENPA overexpression promotes genome instability in pRb-depleted human cells
Abstract
Background: Aneuploidy is a hallmark of most human cancers that arises as a consequence of chromosomal instability and it is frequently associated with centrosome amplification. Functional inactivation of the Retinoblastoma protein (pRb) has been indicated as a cause promoting chromosomal instability as well centrosome amplification. However, the underlying molecular mechanism still remains to be clarified.
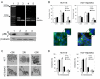
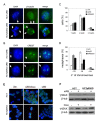
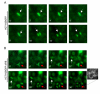
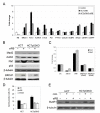
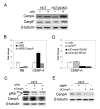
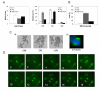
Results: Here we show that pRb depletion both in wild type and p53 knockout HCT116 cells was associated with the presence of multipolar spindles, anaphase bridges, lagging chromosomes and micronuclei harbouring whole chromosomes. In addition aneuploidy caused by pRb acute loss was not affected by p53 loss.Quantitative real-time RT-PCR showed that pRB depletion altered expression of genes involved in centrosome duplication, kinetochore assembly and in the Spindle Assembly Checkpoint (SAC). However, despite MAD2 up-regulation pRb-depleted cells seemed to have a functional SAC since they arrested in mitosis after treatments with mitotic poisons. Moreover pRb-depleted HCT116 cells showed BRCA1 overexpression that seemed responsible for MAD2 up-regulation.Post-transcriptional silencing of CENPA by RNA interference, resulting in CENP-A protein levels similar to those present in control cells greatly reduced aneuploid cell numbers in pRb-depleted cells.
Conclusion: Altogether our findings indicate a novel aspect of pRb acute loss that promotes aneuploidy mainly by inducing CENPA overexpression that in turn might induce micronuclei by affecting the correct attachment of spindle microtubules to kinetochores.
Figures
References
-
- Gimenez-Abian JF, Diaz-Martinez LA, Wirth KG, Andrews CA, Gimenez-Martin G, Clarke DJ. Regulated separation of sister centromeres depends on the spindle assembly checkpoint but not on the anaphase promoting complex/cyclosome. Cell Cycle. 2005;4:1561–1575. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

