The low-affinity phosphate transporter PitA is dispensable for in vitro growth of Mycobacterium smegmatis
- PMID: 20003273
- PMCID: PMC2797804
- DOI: 10.1186/1471-2180-9-254
The low-affinity phosphate transporter PitA is dispensable for in vitro growth of Mycobacterium smegmatis
Abstract
Background: Mycobacteria have been shown to contain an apparent redundancy of high-affinity phosphate uptake systems, with two to four copies of such systems encoded in all mycobacterial genomes sequenced to date. In addition, all mycobacteria also contain at least one gene encoding the low-affinity phosphate transporter, Pit. No information is available on a Pit system from a Gram-positive microorganism, and the importance of this system in a background of multiple other phosphate transporters is unclear.
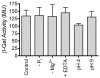
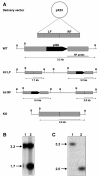
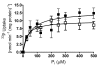
Results: The aim of this study was to determine the physiological role of the PitA phosphate transporter in Mycobacterium smegmatis. Expression of pitA was found to be constitutive under a variety of growth conditions. An unmarked deletion mutant in pitA of M. smegmatis was created. The deletion did not affect in vitro growth or phosphate uptake of M. smegmatis. Expression of the high-affinity transporters, PstSCAB and PhnDCE, was increased in the pitA deletion strain.
Conclusion: PitA is the only low-affinity phosphate transport system annotated in the genome of M. smegmatis. The lack of phenotype of the pitA deletion strain shows that this system is dispensable for in vitro growth of this organism. However, increased expression of the remaining phosphate transporters in the mutant indicates a compensatory mechanism and implies that PitA is indeed used for the uptake of phosphate in M. smegmatis.
Figures
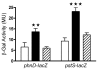
Similar articles
-
Differential regulation of high-affinity phosphate transport systems of Mycobacterium smegmatis: identification of PhnF, a repressor of the phnDCE operon.J Bacteriol. 2008 Feb;190(4):1335-43. doi: 10.1128/JB.01764-07. Epub 2007 Dec 14. J Bacteriol. 2008. PMID: 18083811 Free PMC article.
-
The Phn system of Mycobacterium smegmatis: a second high-affinity ABC-transporter for phosphate.Microbiology (Reading). 2006 Nov;152(Pt 11):3453-3465. doi: 10.1099/mic.0.29201-0. Microbiology (Reading). 2006. PMID: 17074913
-
Deletion of SenX3-RegX3, a key two-component regulatory system of Mycobacterium smegmatis, results in growth defects under phosphate-limiting conditions.Microbiology (Reading). 2012 Nov;158(Pt 11):2724-2731. doi: 10.1099/mic.0.060319-0. Epub 2012 Sep 6. Microbiology (Reading). 2012. PMID: 22956756
-
Molecular Mechanisms of Phosphate Sensing, Transport and Signalling in Streptomyces and Related Actinobacteria.Int J Mol Sci. 2021 Jan 23;22(3):1129. doi: 10.3390/ijms22031129. Int J Mol Sci. 2021. PMID: 33498785 Free PMC article. Review.
-
Nutrient acquisition by mycobacteria.Microbiology (Reading). 2008 Mar;154(Pt 3):679-692. doi: 10.1099/mic.0.2007/012872-0. Microbiology (Reading). 2008. PMID: 18310015 Review.
Cited by
-
Marine picocyanobacterial PhnD1 shows specificity for various phosphorus sources but likely represents a constitutive inorganic phosphate transporter.ISME J. 2023 Jul;17(7):1040-1051. doi: 10.1038/s41396-023-01417-w. Epub 2023 Apr 22. ISME J. 2023. PMID: 37087502 Free PMC article.
-
Genomic Analysis of Pseudomonas sp. Strain SCT, an Iodate-Reducing Bacterium Isolated from Marine Sediment, Reveals a Possible Use for Bioremediation.G3 (Bethesda). 2019 May 7;9(5):1321-1329. doi: 10.1534/g3.118.200978. G3 (Bethesda). 2019. PMID: 30910818 Free PMC article.
-
Regulatory mechanism of the SenX3-RegX3 two-component system in Mycobacterium smegmatis: Roles of PhoU in sensing inorganic phosphate levels.J Biol Chem. 2025 Jun 28;301(8):110435. doi: 10.1016/j.jbc.2025.110435. Online ahead of print. J Biol Chem. 2025. PMID: 40588129 Free PMC article.
-
The complete genome sequence of the plant growth-promoting bacterium Pseudomonas sp. UW4.PLoS One. 2013;8(3):e58640. doi: 10.1371/journal.pone.0058640. Epub 2013 Mar 13. PLoS One. 2013. PMID: 23516524 Free PMC article.
-
Phosphite synthetic auxotrophy as an effective biocontainment strategy for the industrial chassis Pseudomonas putida.Microb Cell Fact. 2022 Aug 8;21(1):156. doi: 10.1186/s12934-022-01883-5. Microb Cell Fact. 2022. PMID: 35934698 Free PMC article.
References
-
- Wanner BL. In: Escherichia coli and Salmonella: cellular and molecular biology. 2. Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editor. Vol. 1. Washington, DC: ASM Press; 1996. Phosphorus Assimilation and Control of the Phosphate Regulon; pp. 1357–1381.
-
- van Veen HW, Abee T, Kortstee GJ, Konings WN, Zehnder AJ. Mechanism and energetics of the secondary phosphate transport system of Acinetobacter johnsonii 210A. J Biol Chem. 1993;268(26):19377–19383. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

