An improved empirical bayes approach to estimating differential gene expression in microarray time-course data: BETR (Bayesian Estimation of Temporal Regulation)
- PMID: 20003283
- PMCID: PMC2801687
- DOI: 10.1186/1471-2105-10-409
An improved empirical bayes approach to estimating differential gene expression in microarray time-course data: BETR (Bayesian Estimation of Temporal Regulation)
Abstract
Background: Microarray gene expression time-course experiments provide the opportunity to observe the evolution of transcriptional programs that cells use to respond to internal and external stimuli. Most commonly used methods for identifying differentially expressed genes treat each time point as independent and ignore important correlations, including those within samples and between sampling times. Therefore they do not make full use of the information intrinsic to the data, leading to a loss of power.
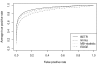
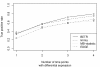
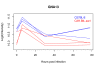
Results: We present a flexible random-effects model that takes such correlations into account, improving our ability to detect genes that have sustained differential expression over more than one time point. By modeling the joint distribution of the samples that have been profiled across all time points, we gain sensitivity compared to a marginal analysis that examines each time point in isolation. We assign each gene a probability of differential expression using an empirical Bayes approach that reduces the effective number of parameters to be estimated.
Conclusions: Based on results from theory, simulated data, and application to the genomic data presented here, we show that BETR has increased power to detect subtle differential expression in time-series data. The open-source R package betr is available through Bioconductor. BETR has also been incorporated in the freely-available, open-source MeV software tool available from http://www.tm4.org/mev.html.
Figures
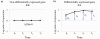
References
-
- Kerr MK, Churchill GA. Statistical design and the analysis of gene expression microarray data. Genet Res. 2001;77(2):123–8. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

