The effects of HIF-1alpha on gene expression profiles of NCI-H446 human small cell lung cancer cells
- PMID: 20003295
- PMCID: PMC2797501
- DOI: 10.1186/1756-9966-28-150
The effects of HIF-1alpha on gene expression profiles of NCI-H446 human small cell lung cancer cells
Abstract
Background: Gene targeted therapy refers to any therapy focused on one of the many biological features of the tumor. Such features are mediated by specific genes that are involved in tumor metastasis, recurrence, poor response to chemotherapy and others. Hypoxia is an important pathognomonic feature of many malignant tumors including SCLC (small cell lung cancer). HIF-1alpha, which is induced by hypoxia, is the most important regulatory factor of many specific genes that can influence the biological features of tumors.
Methods: In this study, we tried to elucidate the changes in gene expression profiles of SCLC NCI-H446 cells mediated by HIF-1alpha. According to different treatments of cells, three experimental pairwise comparisons were designed: hypoxia group vs. control group, Ad5-HIF-1alpha group vs. Ad5 group, and Ad5-siHIF-1 alpha group Vs Ad5 group.
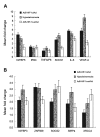
Results: Results from the analysis of gene expression profiles indicated that there were 65 genes upregulated and 28 genes downregulated more than two-fold in all three experimental pairwise comparisons. These genes were involved in transport, signal-transduction, cell adhesion/motility, growth factor/cytokines, transcription, inflammatory response, metabolic process, in addition to others. SOCS1, IGFBP5, IL-6 and STAT3 were also upregulated at protein level. SOCS1 could significantly induce apoptosis and suppress growth of NCI-H446 cells but HIF-1alpha could induce growth and suppress apoptosis.
Conclusions: Through this research, we are trying to find novel functional genes that are mediated by HIF-1alpha and provide the theoretical basis for new therapeutic targets. HIF-1 alpha maybe upregulate the expression of SOCS1 through mediation of STAT3 and IL-6. In addition, SOCS1 could significantly induce apoptosis and suppress growth of NCI-H446 cells. This was contrary to HIF-1alpha and it indicated that there might be an antagonism effect between HIF-1alpha and SOCS1 on regulating growth and apoptosis of NCI-H446 cells.
Figures


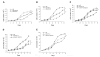
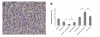
References
-
- Vaupel P, Kallinowski F, Okunieff P. Blood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: a review. Cancer Res. 1989;49:6449–6465. - PubMed
-
- Fan LF, Diao LM. Effect of Hypoxia Induced Factor-1a on the Growth of A549 lung cancer cells. J Pract Med. 2007;23:451–453.
-
- Maxwell PH, Pugh CW, Ratcliffe PJ. Activation of the HIF pathway in cancer. Curr Opin Genet Dev. 2001;11:293–299. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

