Expansion of tandem repeats in sea anemone Nematostella vectensis proteome: A source for gene novelty?
- PMID: 20003297
- PMCID: PMC2805694
- DOI: 10.1186/1471-2164-10-593
Expansion of tandem repeats in sea anemone Nematostella vectensis proteome: A source for gene novelty?
Abstract
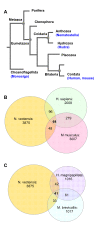
Background: The complete proteome of the starlet sea anemone, Nematostella vectensis, provides insights into gene invention dating back to the Cnidarian-Bilaterian ancestor. With the addition of the complete proteomes of Hydra magnipapillata and Monosiga brevicollis, the investigation of proteins having unique features in early metazoan life has become practical. We focused on the properties and the evolutionary trends of tandem repeat (TR) sequences in Cnidaria proteomes.
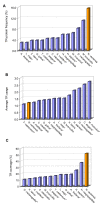
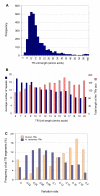
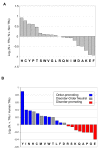
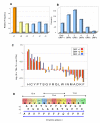
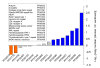
Results: We found that 11-16% of N. vectensis proteins contain tandem repeats. Most TRs cover 150 amino acid segments that are comprised of basic units of 5-20 amino acids. In total, the N. Vectensis proteome has about 3300 unique TR-units, but only a small fraction of them are shared with H. magnipapillata, M. brevicollis, or mammalian proteomes. The overall abundance of these TRs stands out relative to that of 14 proteomes representing the diversity among eukaryotes and within the metazoan world. TR-units are characterized by a unique composition of amino acids, with cysteine and histidine being over-represented. Structurally, most TR-segments are associated with coiled and disordered regions. Interestingly, 80% of the TR-segments can be read in more than one open reading frame. For over 100 of them, translation of the alternative frames would result in long proteins. Most domain families that are characterized as repeats in eukaryotes are found in the TR-proteomes from Nematostella and Hydra.
Conclusions: While most TR-proteins have originated from prediction tools and are still awaiting experimental validations, supportive evidence exists for hundreds of TR-units in Nematostella. The existence of TR-proteins in early metazoan life may have served as a robust mode for novel genes with previously overlooked structural and functional characteristics.
Figures
Similar articles
-
Short toxin-like proteins abound in Cnidaria genomes.Toxins (Basel). 2012 Nov 16;4(11):1367-84. doi: 10.3390/toxins4111367. Toxins (Basel). 2012. PMID: 23202321 Free PMC article.
-
A diverse array of creatine kinase and arginine kinase isoform genes is present in the starlet sea anemone Nematostella vectensis, a cnidarian model system for studying developmental evolution.Gene. 2012 Apr 15;497(2):214-27. doi: 10.1016/j.gene.2012.01.036. Epub 2012 Jan 28. Gene. 2012. PMID: 22305986
-
Rising starlet: the starlet sea anemone, Nematostella vectensis.Bioessays. 2005 Feb;27(2):211-21. doi: 10.1002/bies.20181. Bioessays. 2005. PMID: 15666346 Review.
-
Nuclear receptor complement of the cnidarian Nematostella vectensis: phylogenetic relationships and developmental expression patterns.BMC Evol Biol. 2009 Sep 10;9:230. doi: 10.1186/1471-2148-9-230. BMC Evol Biol. 2009. PMID: 19744329 Free PMC article.
-
Environmental sensing and response genes in cnidaria: the chemical defensome in the sea anemone Nematostella vectensis.Cell Biol Toxicol. 2008 Dec;24(6):483-502. doi: 10.1007/s10565-008-9107-5. Epub 2008 Oct 28. Cell Biol Toxicol. 2008. PMID: 18956243 Free PMC article. Review.
Cited by
-
Short toxin-like proteins abound in Cnidaria genomes.Toxins (Basel). 2012 Nov 16;4(11):1367-84. doi: 10.3390/toxins4111367. Toxins (Basel). 2012. PMID: 23202321 Free PMC article.
-
A haplotype resolved chromosomal level avocado genome allows analysis of novel avocado genes.Hortic Res. 2022 Mar 30;9:uhac157. doi: 10.1093/hr/uhac157. eCollection 2022. Hortic Res. 2022. PMID: 36204209 Free PMC article.
-
Genetic diversity of the allodeterminant alr2 in Hydractinia symbiolongicarpus.Mol Biol Evol. 2011 Feb;28(2):933-47. doi: 10.1093/molbev/msq282. Epub 2010 Oct 21. Mol Biol Evol. 2011. PMID: 20966116 Free PMC article.
References
-
- Alba MM, Tompa P, Veitia RA. Amino acid repeats and the structure and evolution of proteins. Genome Dyn. 2007;3:119–130. full_text. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

