Promoter methylation of IGFBP-3 and p53 expression in ovarian endometrioid carcinoma
- PMID: 20003326
- PMCID: PMC2799391
- DOI: 10.1186/1476-4598-8-120
Promoter methylation of IGFBP-3 and p53 expression in ovarian endometrioid carcinoma
Abstract
Background: Insulin-like growth factor binding protein (IGFBP-3) is an antiproliferative, pro-apoptotic and invasion suppressor protein which is transcriptionally regulated by p53. Promoter methylation has been linked to gene silencing and cancer progression. We studied the correlation between IGFBP-3 and p53 expression as well as IGFBP-3 promoter methylation in ovarian endometrioid carcinoma (OEC) by immunohistochemical staining and quantitative methylation-specific PCR (qMSP). Additionally, we assessed the molecular regulatory mechanism of wild type (wt) p53 on IGFBP-3 expression using two subclones of OEC, the OVTW59-P0 (low invasive) and P4 (high invasive) sublines.
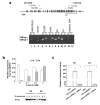
Results: In 60 cases of OEC, 40.0% showed lower IGFBP-3 expression which was significantly correlated with higher IGFBP-3 promoter methylation. p53 overexpression was detected in 35.0% of OEC and was unrelated to clinical outcomes and IGFBP-3. By Kaplan-Meier analysis, patients with lower IGFBP-3, higher IGFBP-3 promoter methylation, and normal p53 were associated most significantly with lower survival rates. In OEC cell line, IGFBP-3 expression was correlated with IGFBP-3 promoter methylation. IGFBP-3 expression was restored after treatment with a DNA methy-transferase inhibitors (5-aza-deoxycytidine) and suppressed by a p53 inhibitor (pifithrin-alpha). The putative p53 regulatory sites on the promoter of IGFBP-3 were identified at -210, -206, -183 and -179 bases upstream of the transcription start site. Directed mutagenesis at these sites quantitatively reduced the transcription activity of IGFBP-3.
Conclusion: Our data suggests that IGFBP-3 silencing through IGFBP-3 promoter methylation in the absence of p53 overexpression is associated with cancer progression. These results support a potential role of IGFBP-3 methylation in the carcinogenesis of OEC.
Figures



Similar articles
-
Insulin-like growth factor binding protein-3 (IGFBP-3) acts as an invasion-metastasis suppressor in ovarian endometrioid carcinoma.Oncogene. 2008 Apr 3;27(15):2137-47. doi: 10.1038/sj.onc.1210864. Epub 2007 Oct 22. Oncogene. 2008. PMID: 17952116
-
Functional promoter upstream p53 regulatory sequence of IGFBP3 that is silenced by tumor specific methylation.BMC Cancer. 2005 Jan 20;5:9. doi: 10.1186/1471-2407-5-9. BMC Cancer. 2005. PMID: 15661074 Free PMC article.
-
Regularly methylated novel pro-apoptotic genes associated with recurrence in transitional cell carcinoma of the bladder.Int J Cancer. 2006 Sep 15;119(6):1396-402. doi: 10.1002/ijc.21971. Int J Cancer. 2006. PMID: 16642478
-
Methylation of the insulin-like growth factor binding protein-3 gene and prognosis of epithelial ovarian cancer.Int J Gynecol Cancer. 2006 Jan-Feb;16(1):210-8. doi: 10.1111/j.1525-1438.2006.00299.x. Int J Gynecol Cancer. 2006. PMID: 16445635
-
Promoter hypermethylation profile of kidney cancer with new proapoptotic p53 target genes and clinical implications.Clin Cancer Res. 2006 Sep 1;12(17):5040-6. doi: 10.1158/1078-0432.CCR-06-0144. Clin Cancer Res. 2006. PMID: 16951219
Cited by
-
Low expression of IGFBP-3 predicts poor prognosis in patients with esophageal squamous cell carcinoma.Med Oncol. 2012 Dec;29(4):2669-76. doi: 10.1007/s12032-011-0133-4. Epub 2011 Dec 14. Med Oncol. 2012. PMID: 22167391
-
Current data and future perspectives on DNA methylation in ovarian cancer (Review).Int J Oncol. 2024 Jun;64(6):62. doi: 10.3892/ijo.2024.5650. Epub 2024 May 17. Int J Oncol. 2024. PMID: 38757340 Free PMC article. Review.
-
Cellular and clinical impact of protein phosphatase enzyme epigenetic silencing in multiple cancer tissues.Hum Genomics. 2024 Mar 12;18(1):24. doi: 10.1186/s40246-024-00592-x. Hum Genomics. 2024. PMID: 38475971 Free PMC article.
-
Epigenetic regulation of insulin-like growth factor binding protein-3 (IGFBP-3) in cancer.J Cell Commun Signal. 2015 Jun;9(2):159-66. doi: 10.1007/s12079-015-0294-6. Epub 2015 Apr 29. J Cell Commun Signal. 2015. PMID: 25920743 Free PMC article.
-
Molecular epigenetics in the management of ovarian cancer: are we investigating a rational clinical promise?Front Oncol. 2014 Apr 8;4:71. doi: 10.3389/fonc.2014.00071. eCollection 2014. Front Oncol. 2014. PMID: 24782983 Free PMC article. Review.
References
-
- McGuire WP, Hoskins WJ, Brady MF, Kucera PR, Partridge EE, Look KY, Clarke-Pearson DL, Davidson M. Cyclophosphamide and cisplatin versus paclitaxel and cisplatin: a phase III randomized trial in patients with suboptimal stage III/IV ovarian cancer (from the Gynecologic Oncology Group) Semin Onco. 1996;23:40–47. - PubMed
-
- Bhattacharyya N, Pechhold K, Shahjee H, Zappala G, Elbi C, Raaka B, Wiench M, Hong J, Rechler MM. Nonsecreted insulin-like growth factor binding protein-3 (IGFBP-3) can induce apoptosis in human prostate cancer cells by IGF-independent mechanisms without being concentrated in the nucleus. J Biol Chem. 2006;281:24588–24601. doi: 10.1074/jbc.M509463200. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

