The transcriptional programme of Salmonella enterica serovar Typhimurium reveals a key role for tryptophan metabolism in biofilms
- PMID: 20003355
- PMCID: PMC2805695
- DOI: 10.1186/1471-2164-10-599
The transcriptional programme of Salmonella enterica serovar Typhimurium reveals a key role for tryptophan metabolism in biofilms
Abstract
Background: Biofilm formation enhances the capacity of pathogenic Salmonella bacteria to survive stresses that are commonly encountered within food processing and during host infection. The persistence of Salmonella within the food chain has become a major health concern, as biofilms can serve as a reservoir for the contamination of food products. While the molecular mechanisms required for the survival of bacteria on surfaces are not fully understood, transcriptional studies of other bacteria have demonstrated that biofilm growth triggers the expression of specific sets of genes, compared with planktonic cells. Until now, most gene expression studies of Salmonella have focused on the effect of infection-relevant stressors on virulence or the comparison of mutant and wild-type bacteria. However little is known about the physiological responses taking place inside a Salmonella biofilm.
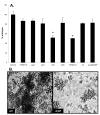
Results: We have determined the transcriptomic and proteomic profiles of biofilms of Salmonella enterica serovar Typhimurium. We discovered that 124 detectable proteins were differentially expressed in the biofilm compared with planktonic cells, and that 10% of the S. Typhimurium genome (433 genes) showed a 2-fold or more change in the biofilm compared with planktonic cells. The genes that were significantly up-regulated implicated certain cellular processes in biofilm development including amino acid metabolism, cell motility, global regulation and tolerance to stress. We found that the most highly down-regulated genes in the biofilm were located on Salmonella Pathogenicity Island 2 (SPI2), and that a functional SPI2 secretion system regulator (ssrA) was required for S. Typhimurium biofilm formation. We identified STM0341 as a gene of unknown function that was needed for biofilm growth. Genes involved in tryptophan (trp) biosynthesis and transport were up-regulated in the biofilm. Deletion of trpE led to decreased bacterial attachment and this biofilm defect was restored by exogenous tryptophan or indole.
Conclusions: Biofilm growth of S. Typhimurium causes distinct changes in gene and protein expression. Our results show that aromatic amino acids make an important contribution to biofilm formation and reveal a link between SPI2 expression and surface-associated growth in S. Typhimurium.
Figures




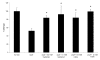
References
-
- Helke DM, Wong ACL. Survival and growth characteristics of Listeria monocytogenes and Salmonella Typhimurium on stainless steel and buna-n-rubber. J Food Prot. 1994;57:963–968. http://www.ingentaconnect.com/content/iafp/jfp/1994/00000057/00000011/ar... - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

