TIM-Finder: a new method for identifying TIM-barrel proteins
- PMID: 20003393
- PMCID: PMC2803183
- DOI: 10.1186/1472-6807-9-73
TIM-Finder: a new method for identifying TIM-barrel proteins
Abstract
Background: The triosephosphate isomerase (TIM)-barrel fold occurs frequently in the proteomes of different organisms, and the known TIM-barrel proteins have been found to play diverse functional roles. To accelerate the exploration of the sequence-structure protein landscape in the TIM-barrel fold, a computational tool that allows sensitive detection of TIM-barrel proteins is required.
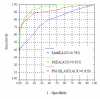
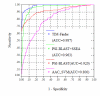
Results: To develop a new TIM-barrel protein identification method in this work, we consider three descriptors: a sequence-alignment-based descriptor using PSI-BLAST e-values and bit scores, a descriptor based on secondary structure element alignment (SSEA), and a descriptor based on the occurrence of PROSITE functional motifs. With the assistance of Support Vector Machine (SVM), the three descriptors were combined to obtain a new method with improved performance, which we call TIM-Finder. When tested on the whole proteome of Bacillus subtilis, TIM-Finder is able to detect 194 TIM-barrel proteins at a 99% confidence level, outperforming the PSI-BLAST search as well as one existing fold recognition method.
Conclusions: TIM-Finder can serve as a competitive tool for proteome-wide TIM-barrel protein identification. The TIM-Finder web server is freely accessible at http://202.112.170.199/TIM-Finder/.
Figures


Similar articles
-
DescFold: a web server for protein fold recognition.BMC Bioinformatics. 2009 Dec 14;10:416. doi: 10.1186/1471-2105-10-416. BMC Bioinformatics. 2009. PMID: 20003426 Free PMC article.
-
Descriptor-based protein remote homology identification.Protein Sci. 2005 Feb;14(2):431-44. doi: 10.1110/ps.041035505. Epub 2005 Jan 4. Protein Sci. 2005. PMID: 15632283 Free PMC article.
-
The TIM Barrel Architecture Facilitated the Early Evolution of Protein-Mediated Metabolism.J Mol Evol. 2016 Jan;82(1):17-26. doi: 10.1007/s00239-015-9722-8. Epub 2016 Jan 5. J Mol Evol. 2016. PMID: 26733481 Free PMC article.
-
The TIM-barrel fold: a versatile framework for efficient enzymes.FEBS Lett. 2001 Mar 16;492(3):193-8. doi: 10.1016/s0014-5793(01)02236-0. FEBS Lett. 2001. PMID: 11257493 Review.
-
A guide to the effects of a large portion of the residues of triosephosphate isomerase on catalysis, stability, druggability, and human disease.Proteins. 2017 Jul;85(7):1190-1211. doi: 10.1002/prot.25299. Epub 2017 Apr 27. Proteins. 2017. PMID: 28378917 Review.
Cited by
-
Insights into the fold organization of TIM barrel from interaction energy based structure networks.PLoS Comput Biol. 2012;8(5):e1002505. doi: 10.1371/journal.pcbi.1002505. Epub 2012 May 17. PLoS Comput Biol. 2012. PMID: 22615547 Free PMC article.
-
Computational Prediction of RNA-Binding Proteins and Binding Sites.Int J Mol Sci. 2015 Nov 3;16(11):26303-17. doi: 10.3390/ijms161125952. Int J Mol Sci. 2015. PMID: 26540053 Free PMC article. Review.
-
Recognition of Protein Pupylation Sites by Adopting Resampling Approach.Molecules. 2018 Nov 27;23(12):3097. doi: 10.3390/molecules23123097. Molecules. 2018. PMID: 30486421 Free PMC article.
-
ET-GRU: using multi-layer gated recurrent units to identify electron transport proteins.BMC Bioinformatics. 2019 Jul 6;20(1):377. doi: 10.1186/s12859-019-2972-5. BMC Bioinformatics. 2019. PMID: 31277574 Free PMC article.
-
Prediction of ubiquitination sites by using the composition of k-spaced amino acid pairs.PLoS One. 2011;6(7):e22930. doi: 10.1371/journal.pone.0022930. Epub 2011 Jul 29. PLoS One. 2011. PMID: 21829559 Free PMC article.
References
-
- Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP:a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247(4):536–540. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

