Defining the healthy "core microbiome" of oral microbial communities
- PMID: 20003481
- PMCID: PMC2805672
- DOI: 10.1186/1471-2180-9-259
Defining the healthy "core microbiome" of oral microbial communities
Abstract
Background: Most studies examining the commensal human oral microbiome are focused on disease or are limited in methodology. In order to diagnose and treat diseases at an early and reversible stage an in-depth definition of health is indispensible. The aim of this study therefore was to define the healthy oral microbiome using recent advances in sequencing technology (454 pyrosequencing).
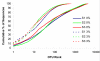
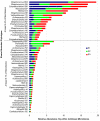
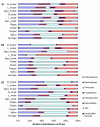
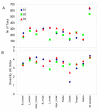
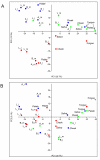
Results: We sampled and sequenced microbiomes from several intraoral niches (dental surfaces, cheek, hard palate, tongue and saliva) in three healthy individuals. Within an individual oral cavity, we found over 3600 unique sequences, over 500 different OTUs or "species-level" phylotypes (sequences that clustered at 3% genetic difference) and 88 - 104 higher taxa (genus or more inclusive taxon). The predominant taxa belonged to Firmicutes (genus Streptococcus, family Veillonellaceae, genus Granulicatella), Proteobacteria (genus Neisseria, Haemophilus), Actinobacteria (genus Corynebacterium, Rothia, Actinomyces), Bacteroidetes (genus Prevotella, Capnocytophaga, Porphyromonas) and Fusobacteria (genus Fusobacterium).Each individual sample harboured on average 266 "species-level" phylotypes (SD 67; range 123 - 326) with cheek samples being the least diverse and the dental samples from approximal surfaces showing the highest diversity. Principal component analysis discriminated the profiles of the samples originating from shedding surfaces (mucosa of tongue, cheek and palate) from the samples that were obtained from solid surfaces (teeth).There was a large overlap in the higher taxa, "species-level" phylotypes and unique sequences among the three microbiomes: 84% of the higher taxa, 75% of the OTUs and 65% of the unique sequences were present in at least two of the three microbiomes. The three individuals shared 1660 of 6315 unique sequences. These 1660 sequences (the "core microbiome") contributed 66% of the reads. The overlapping OTUs contributed to 94% of the reads, while nearly all reads (99.8%) belonged to the shared higher taxa.
Conclusions: We obtained the first insight into the diversity and uniqueness of individual oral microbiomes at a resolution of next-generation sequencing. We showed that a major proportion of bacterial sequences of unrelated healthy individuals is identical, supporting the concept of a core microbiome at health.
Figures


References
-
- Wilson M. Bacteriology of Humans: An Ecological Perspective. Malden, MA: Blackwell Publishing Ltd; 2008.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

